Abstract
Heterotrimeric G proteins (αβγ) mediate the majority of signaling pathways in mammalian cells. It is long held that G protein function is localized to the plasma membrane. Here we examined the spatiotemporal dynamics of G protein localization using fluorescence recovery after photobleaching, fluorescence loss in photobleaching, and a photoswitchable fluorescent protein, Dronpa. Unexpectedly, G protein subunits shuttle rapidly (t½ < 1 min) between the plasma membrane and intracellular membranes. We show that consistent with such shuttling, G proteins constitutively reside in endomembranes. Furthermore, we show that shuttling is inhibited by 2-bromopalmitate. Thus, contrary to present thought, G proteins do not reside permanently on the plasma membrane but are constantly testing the cytoplasmic surfaces of the plasma membrane and endomembranes to maintain G protein pools in intracellular membranes to establish direct communication between receptors and endomembranes.
Heterotrimeric G proteins are known to function on the cytosolic surface of the plasma membrane (PM)2 in response to the stimulation of transmembrane receptors by most of the extracellular signals a mammalian cell senses (1–4). The PM localization of a G protein is facilitated by individual lipid modifications of the α and βγ subunits (5). Consistent with a model of G protein action on the PM, activated G protein subunits have been shown to modulate the functions of effectors such as adenylyl cyclase, phospholipase C, and ion channels which are also PM-localized (1–4). However, the spatiotemporal dynamics of G protein localization in a living cell has not been examined to determine whether G protein subunits are stably constrained to the PM. Although there is longstanding evidence for the existence of G protein subunits in intracellular membranes such as the Golgi complex and suggestions that they affect protein trafficking (6–11), it has been unclear as to how they reach endomembranes and if they are native to these membranes or in transit to the PM.
Here we used a variety of imaging methods on live cells with fluorescent protein-tagged G protein subunits including photoswitchable Dronpa (12) to observe G protein movement. A fluorescence resonance energy transfer (FRET)-based G protein sensor was used to examine whether G proteins reside constitutively in the endomembranes. The results show that G protein subunits shuttle rapidly between the PM and endomembranes in cells in the basal state maintaining a pool of G proteins in the endomembranes. We show that the shuttling is likely diffusive and not vesicle-mediated. Furthermore, 2-bromopalmitate (2BP), an inhibitor of palmitoylation (13), inhibited shuttling, suggesting that it may be regulated by acylation.
MATERIALS AND METHODS
Details of chemicals, expression constructs, cell lines, transfection, and treatment of cells are as in Saini et al. (41). Dronpa fluorescent proteins were introduced downstream of Gly-92 in αo as in the αo-CFP construct that we have shown previously topossess normal functional properties (14) or at the N terminus of γ subunits.
Image Acquisition and Processing
Cells were cultured in 35-mm glass bottom dishes (World Precision Instruments) or on acid-washed glass coverslips and transiently transfected with appropriate combinations of different G protein subunits as described in the text and figure legends. After 16–24 h post-transfection the cells were processed for imaging. Cells were washed with Hanks’ buffer saline solution supplemented with 10 mM HEPES, pH 7.4, and maintained in this buffer during imaging experiments. For FRET experiments, the coverslips were mounted on an imaging chamber with an internal volume of 25 μl (Warner Instruments). Details are available in Saini et al. (41).
For confocal microscopy cells were visualized with an Olympus LSM Fluoview FV300 microscope using a 40× oil immersion objective (1.3 NA). Images of cells expressing YFP, Dronpa, and green fluorescent protein (GFP) were acquired with a Multi-line argon laser using 488-nm excitation and with emission filter BA 505–525. Images of galactosyl transferase-DsRed monomer were acquired using a Green HeNe laser line for excitation at 543 nm with emission filter BA 560–600. Images were acquired with Fluoview FV300 software and then processed with MetaMorph software (Molecular Devices Corp., Downingtown, PA). For FRAP, FLIP, and analysis using Dronpa, images were acquired with the lasers mentioned above. Laser intensity for YFP/GFP/Dronpa excitation was 0.5–1% (488 nm) and for Gal-T-DsRed-monomer excitation was 5–10% (543 nm). To restrict UV damage in Dronpa photoactivation (PA) experiments, selective PA of the Golgi was performed at 458 nm (100% intensity) for 2–3 s after whole cell bleaching (488 nm at 100% laser intensity). In FRAP and FLIP experiments Golgi or cytosolic region was bleached for 1–2 s at 488 nm with 100% laser intensity. All bleaching and photoactivation was done using iterative scanning (10 times for bleaching and 20 times for PA). The first point in the FRAP and PA plots includes the time for bleaching or PA and confocal lag time, which was constant at 4 s.
All FRAP experiments were performed with transiently transfected cells except for γ11, which was analyzed using previously characterized CHO cells stably expressing muscarinic M2 receptor (M2-CHO) (15) and G protein subunits αo-CFP, β1, and YFP-γ11. Image brightness and contrast were altered equally for the entire series of images using Metamorph to allow better visualization of the recovery in the Golgi complex. Intrinsic pixel intensity values were not altered by these changes.
For FRET analysis through photobleaching experiments, the FRET was determined by monitoring gain in CFP emission intensity in PM or Golgi by photobleaching of YFP (acceptor photobleaching) (14). Cells with equal expression levels of CFP and YFP were selected. CFP and YFP images were acquired. YFP was then photobleached for 2–3 min using YFP excitation without any neutral density filter. After photobleaching of YFP, another CFP image was captured and subsequently CFP intensity from PM or Golgi was analyzed from pre- and post-photobleaching images for calculating FRET efficiency. Percentage change in CFP signal was calculated using the formula (CCfinal − CCinitial/CCinitial) × 100, where CC is CFP emission intensity. Initial and final values were those before and after photobleaching. The emission intensity was corrected for CFP bleaching by determining it in cells expressing αo-CFP alone. In agonist-treated cells, the cells were first stimulated with 100 μM carbachol, and FRET was examined in the same way as described above.
RESULTS
G Protein Subunits Shuttle between PM and Endomembranes
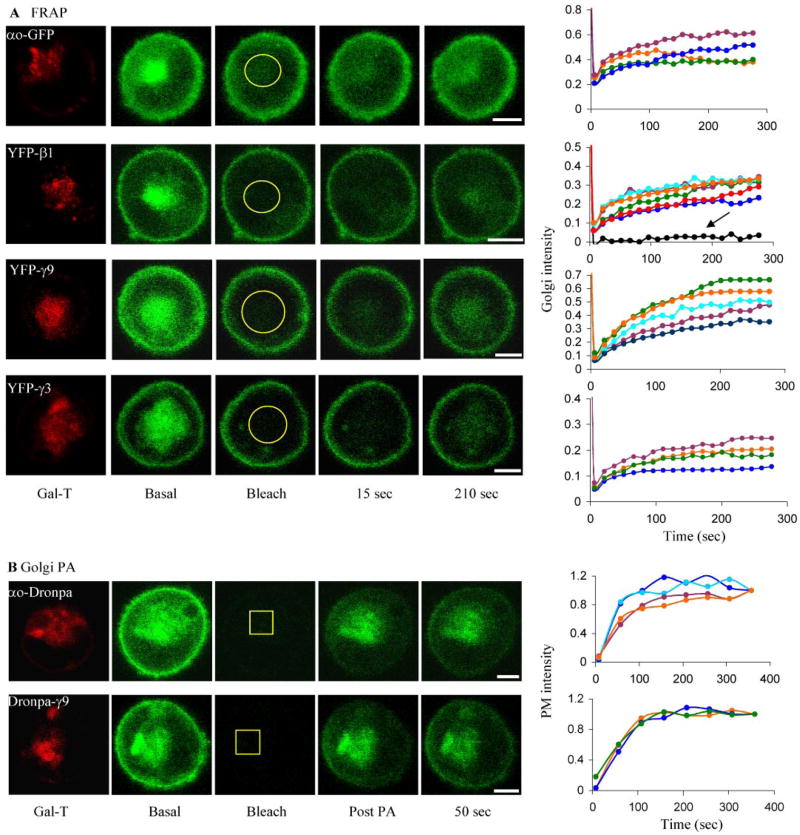
To explore the spatiotemporal dynamics of G protein subunit localization in a live cell, we expressed the αo subunit tagged with either GFP or Dronpa, a photoswitchable fluorescent protein (12), in CHO cells. Fluorescent proteins were independently introduced downstream of Gly-92 in αo as in the αo-CFP construct that we have shown previously to possess normal functional properties (14). Apart from the PM, αo-GFP was localized to the endomembranes, predominantly to the Golgi complex based on colocalization with a Golgi marker (galactosyl transferase (GalT)) (Fig. 1). When Golgi fluorescence was bleached and the recovery in the bleached region was determined (FRAP), fluorescence recovered rapidly, indicating retrograde movement (Fig. 1A). The recovery seen indicated a t½ less than 1 min. We similarly examined a G protein β subunit, β1, the predominant β subunit in mammalian tissues. Similar to the α subunit, it was distributed in the PM and in endomembranes, predominantly Golgi (Fig. 1A). The resolutions of the images that we have obtained do not preclude the possibility that some of the signal from the tagged G protein subunits in the PM is from endosomes in proximity to the PM. In FRAP experiments fluorescence recovered rapidly in the Golgi region of cells expressing YFP-β1 (Fig. 1A). The recovery seen with the fluorescent protein-tagged G protein subunits in the FRAP experiments could not be ascribed to intrinsic recovery of GFP or YFP fluorescence or new protein synthesis because it was <5% in cells that were fully bleached over the period of the experiment (e.g. Fig. 1A, right, black line in YFP-β1). Because β subunits are tightly bound to a γ subunit (16), these results also indicated that γ subunits in the cell capable of binding β1 were also shuttling. We then examined different γ subunit types with distinct receptor-induced translocation properties (41). γ9 and γ11 translocate rapidly from the PM to endomembranes on receptor activation, whereas γ2 and γ3 do not translocate. FRAP experiments showed that all four subunits, however, shuttle between the PM and endomembranes. The recovery of γ9 and γ3 is shown in Fig. 1A. Fig. 2, right, blue, shows recovery from several different cells for γ11. γ2 also demonstrated recovery in a similar FRAP experiment (supplemental Fig. 1). The recovery observed in FRAP experiments cannot be ascribed to the movement of fluorescent protein from the Golgi complex in other planes, since the bleaching is complete along the z axis. (supplemental Fig. 2). The recovery of the α, β, and γ subunits was incomplete in each case, consistent with the relatively large size of the Golgi in CHO cells and the high concentration of the tagged subunits in the Golgi of cells examined similar to Ras in Madin-Darby canine kidney cells (17). The partial recovery may also result from the presence of immobile subunits in the membranes.
FIGURE 1. Shuttling of G protein subunits between PM and endomembranes.
Confocal images of CHO cells expressing a Golgi marker galactosyl transferase (GalT)-DsRed-monomer (red) with fluorescent protein-tagged subunits as labeled (green). A, FRAP. The Golgi region identified with galactosyl transferase-DsRed-monomer was bleached (Bleach). Recovery was monitored by acquiring images every 15 s. Representative images at selected times and the corresponding plots of mean pixel intensity in a selected Golgi region (within yellow circle) are shown. Plots showing fluorescence recovery in the endomembranes are normalized to the prebleach intensity value. Recovery is shown for αo-GFP (n = 4); β1 (n = 6), γ9 (n = 5), and γ3 (n = 4). Plots of different colors show recovery in different cells. The second point in the FRAP plots includes the time for bleaching and confocal lag time. Recovery after whole cell photobleaching was measured in YFP-β1 (black line indicated by the arrow) as a control. Bar, 5 μm. B, Golgi PA. Dronpa-tagged αo (n = 4) and γ9 (n = 3); the Golgi region within the yellow square was selected and photoactivated at 458 nm (to restrict UV damage). Images were acquired immediately (post PA) and subsequently every 50 s. Bar, 5 μm. The longer time interval compared with FRAP experiments reduced Dronpa bleaching. The PM recovery is shown in corresponding plots from different cells. Dronpa bleached 5% every time an image was acquired at this laser intensity. Plots were corrected for this loss. The uncorrected plots are shown in supplemental Fig. 3. Plots showing fluorescence recovery in the PM are normalized to the last intensity value. The first point in the PA plots includes the time for PA and confocal lag time.
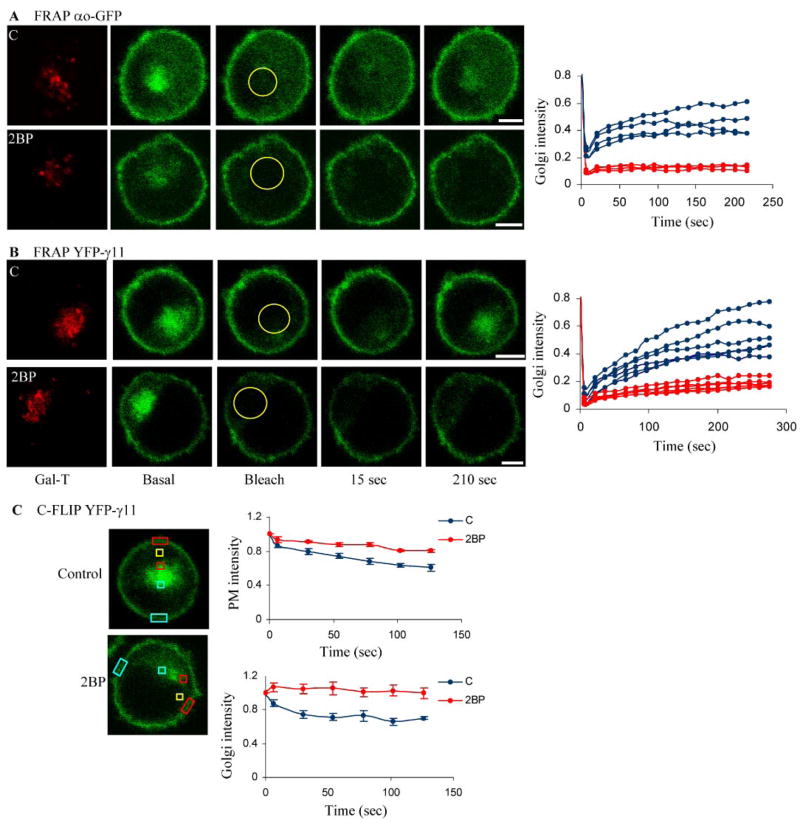
FIGURE 2. Effect of 2BP on G protein subunit shuttling.
Cells were treated with 50 μM 2BP for 30 min at 37 °C. A, FRAP of CHO cells transfected with αo-GFP (n = 4 each) in treated and untreated cells was determined as in Fig. 1. B, FRAP of YFP-γ11 (n = 6 each) stably co-expressed with αo-CFP and β1 in M2-CHO cells. Experimental conditions for FRAP recovery are as in Fig. 1. Plots show changes in fluorescence intensity from individual cells untreated (blue) and treated with 2BP (red). Bar, 5 μm. Yellow circles show the Golgi area monitored for recovery after photobleaching. Plots are normalized to the prebleach intensity value. C, cytosolic FLIP (C-FLIP) of YFP-γ11 stably co-expressed with αo-CFP and β1 in M2-CHO cells. A small area in the cytosol was bleached (yellow square) (6×, 1 s every 15 s). Intensity after each photobleaching event was measured in selected regions on the PM and the Golgi proximal and distal to the bleached region (colored rectangles). In our experimental conditions only 50% of cells treated with 2BP do not show reduction in both proximal PM and Golgi regions, and only the values from those cells are plotted. Plots show the ratio of the mean pixel intensity ± S.E. (n = 5 each) in the proximal/distal regions of the PM (top) and the Golgi (bottom). The values are normalized to the prebleach values. Plots are significantly different at each time point after the second photobleaching in a t test (in the PM, p < 0.01; in the Golgi, p < 0.005).
To test for α subunit anterograde movement, αo-Dronpa was photoactivated in the Golgi (Golgi PA) (Fig. 1B). Fluorescence increased over time in the PM, suggesting rapid movement through the cytosol to the PM. The t½ was <1 min, consistent with the rapidity of the retrograde movement. Golgi PA experiments with Dronpa-γ9 and -γ2 showed that both subunits show anterograde movement similar to αo (Fig. 1B and supplemental Fig. 1). Additional experiments show that the recovery of fluorescence detected in the PM in the Golgi PA experiments is not the result of photoactivation of Dronpa-tagged proteins on the PM above and below the confocal plane. (i) Photoactivation of a relatively small region of the Golgi in Dronpa-γ9-expressing cells results over time in the appearance of fluorescence uniformly over the entire PM, whereas similar PA of a region on the PM results in localized appearance of fluorescence on the PM (supplemental Fig. 4). (ii) Selective bleaching of fluorescence in the PM of a YFP-γ11-expressing cell results in the appearance of fluorescence in the PM over time and a decrease in Golgi region fluorescence (supplemental Fig. 5).
Similar FRAP and Dronpa PA experiments with γ2 showed that this subunit type also shuttles between the PM and endomembranes. Together these results show that regardless of translocation properties, γ subunits shuttle rapidly between the PM and intracellular membranes in the basal state. The ability of βγ2 and βγ3 to shuttle rapidly between PM and Golgi but not translocate on receptor activation is consistent with the prediction that the relative affinities of γ subunits for an activated receptor determine their ability to translocate (18).
Because a mammalian cell in the basal state contains most G proteins in the inactive heterotrimer form so that constitutive activity of the dissociated subunits is curtailed, the shuttling of the different G protein subunits suggests that endogenous α, β, and γ subunits shuttle between the PM and endomembranes. It also suggests that G proteins most likely shuttle as heterotrimers. The shuttling of G protein subunits seen here is not peculiar to CHO cells because both FRAP and PA experiments using a different cell line, HeLa cells, showed rapid shuttling of αo and γ9 between PM and endomembranes (supplemental Fig. 6).
G Protein Shuttling Is Likely Diffusion-mediated
The rapidity of shuttling and the transient appearance of a cytosolic signal in photoactivation experiments suggested that the shuttling of G protein subunits is diffusion-mediated. In addition, the anterograde diffusion of G protein subunits in the basal state and reverse translocation of βγ on receptor inactivation are much faster than the known rate of vesicular transport from Golgi to PM, which is estimated to be 3% per min (19, 20). Consistent with these indicators, nocodazole and monensin, which block vesicular trafficking through different mechanisms (21), did not have any effect on the retrograde or anterograde movement of α or βγ subunits detected using FRAP or Golgi PA (not shown). Supplemental Fig. 7 shows a control for nocodazole action. Rapid retrograde movement of YFP-γ9 occurred at 10 °C. Vesicle-mediated trafficking in cells has been shown to be inhibited at temperatures below 16 °C (22). These results suggest that shuttling between the Golgi and PM is likely to be diffusive.
Shuttling and Translocation Are Inhibited by 2BP
The G protein α subunits belonging to the Gi/o, Gs, and Gq families are palmitoylated (5), and the palmitoyl moiety can be turned over in the basal deactivated state (t½ > 60 min for αi; 90 min for αs) (23–25). We tested whether acylation played a role in the shuttling of G protein subunits and the translocation of the βγ complex between PM and endomembranes by examining the effect of 2BP, an inhibitor that has been used for identifying palmitoylation (13) of proteins. Although 2BP has been used to demonstrate the acylation dependence of protein movement (e.g. Refs. 17 and 26), it is also known to inhibit other processes unrelated to palmitoylation (13, 27). In FRAP experiments 30 min of treatment with 50 μM 2BP inhibited the recovery of fluorescence in the Golgi region of cells expressing αo-GFP, YFP-γ11, or YFP-γ9 (Fig. 2 and supplemental Fig. 8). The variation in inhibition is consistent with previous results using 2BP and is due to the variable effective concentration of 2BP inside cells due to variations in cell density (17, 28). The inhibition of retrograde movement of γ11 occurred in a cell that stably expressed αo-CFP, β1, and YFP-γ11 (Fig. 2B). This result showed that retrograde movement occurs regardless of whether G protein subunits are transiently introduced or stably expressed.
To further examine the G protein subunit movement between the PM and Golgi, cells stably expressing YFP-γ11, β1, and αo-CFP were examined. A region of the cytosol was selected in these cells after scanning along the z axis to ensure lack of overlap with the Golgi and bleached repeatedly (1 s each) with 15-s pauses to allow recovery (cytosolic FLIP, C-FLIP). The resulting images showed a rapid decrease in the proximal but not distal portions of the PM and the Golgi regions (Fig. 2C). When the YFP-γ11 stable cells were treated with 2BP, half the cells examined did not show the differential decrease in fluorescence between the proximal and distal regions of the PM or Golgi seen in untreated cells. The variation in the response to 2BP among cells is consistent with the FRAP results and previous 2BP experiments (17). One interpretation of this behavior of YFP-γ11 is that there is acylation-dependent two-way movement of the protein through the cytosol between the PM and Golgi. Although such rapid shuttling of G proteins from Golgi to PM is unanticipated, it has been previously suggested based on the localization of palmitoyl lacking α subunits that acylation at endomembranes facilitates PM localization of a G protein (29, 30). The 2BP inhibition may also inhibit an unidentified process that is not related to palmitoylation of a G protein subunit or a binding protein (13, 27).
Based on the design of a G protein FRET sensor that we have previously characterized extensively (Fig. 3A, diagram) (14), we stably expressed αo-CFP-β1-YFP-γ11 heterotrimer in CHO cells that also stably express M2 muscarinic receptors. The M2-expressing cells (M2-CHO) have been characterized before (15). A FRET signal was detected in the PM of the cells (Fig. 3A, right, left gray bar). FRET was abrogated on receptor activation with an agonist showing that the G protein present on the PM was capable of getting activated (Fig. 3A, right, left white bar). When the same cells were treated with 2BP, although the relative intensity of YFP and CFP was lower, basal FRET was obtained in 75–80% of the cells (Fig. 3A, right, right gray bar). The FRET was abrogated by agonist addition (Fig. 3A, right, right white bar). This result shows that G protein activation per se is not affected by 2BP treatment.
FIGURE 3. A FRET-based G protein sensor shows G protein activation in 2BP-treated cells and the presence of heterotrimers in the Golgi.

A, in the deactivated heterotrimer FRET occurs from αo-CFP to YFP-γ11. On activation with an agonist, carbachol (100 μM) and G protein subunit dissociation, FRET is abrogated both in control and 2BP-treated cells. FRET efficiency was determined by monitoring gain in CFP emission by photo-bleaching YFP (acceptor). Cells with equal expression levels of CFP and YFP were selected. Results are the means ± S.E. (n = 10 for each condition). Details are under “Materials and Methods” and in supplemental Fig. 9. B, images of M2-CHO cells stably expressing αo-CFP, β1, and YFP-γ11. αo and γ11 are present in the PM and Golgi. The bar diagram shows FRET from the G protein sensor in cells treated with cycloheximide for 6 – 8 h. Results are the means ± S.E. (n = 15).
We notice that the G protein subunits are present in the PM and Golgi after 30 min of 2BP treatment. Similar to the G protein subunits, H-Ras is also present on the PM after 30 min of treatment with 2BP (17, 26). Much longer periods of exposure to 2BP leads to the accumulation of G protein subunits and H-Ras in the Golgi (26, 29). The reasons for these differences are unclear.
G Proteins Are Residents of Endomembranes
There has been evidence for the presence of G protein subunits in endomembranes and suggestions from assays that they may have roles in protein trafficking through the endomembranes and Golgi disassembly (6, 8–11). However, it has been unclear how G protein subunits reach endomembranes and if they are resident therein rather than molecules in transit to the PM through the endomembrane system. Here we detected αo-CFP and YFP-γ11 at approximately equal levels in the Golgi and PM in stably transfected M2-CHO cells (Fig. 3B). Cycloheximide treatment for 6–8 h had no effect on this distribution (not shown). FRET signals of similar efficiencies in the PM and Golgi of cells were detected even in the absence of protein synthesis (Fig. 3B). These results suggest that G proteins exist as heterotrimeric residents of endomembranes.
DISCUSSION
The results here suggest that G proteins shuttle rapidly, continually, and diffusively in the basal state between the PM and endomembranes. The observation here that tagged α, β, and γ subunits shuttle when expressed independently indicates that endogenous G protein subunits in mammalian cells shuttle between the PM and endomembranes. The results also suggest that shuttling occurs in the heterotrimeric form. The G protein heterotrimer is known to be inactive (1–4). Thus, the constant redistribution of G protein molecules occurring between the PM and endomembranes is unlikely to result in changes in downstream signaling activity. However, receptor stimulation allows this shuttling to be harnessed for the translocation of free and potentially active βγ complexes to specific endomembranes at different rates (41).
The rapid shuttling of G protein subunits identified here allows pools of G proteins to coexist in the PM and intracellular membranes in dynamic equilibrium. Proteins diffuse at rates that are at least an order of magnitude more in the cytosol compared with the PM (31). Cytosolic diffusion of G protein subunits thus allows the more rapid movement necessary for constantly sensing the surfaces of internal membranes.
Similar to these unexpected G protein properties, H-Ras and N-Ras have been definitively demonstrated to be functional residents of intracellular membranes and to shuttle between the PM and Golgi controlled by an acylation cycle with t½ values ranging from 1 to 10 min (17, 26, 31–33). Acylation-regulated binding of Ras to the PM was predicted earlier based on the analysis of Ras peptides (34). K-Ras has been shown to translocate on neuronal activation (t½ > 10 min) (35) consistent with the dynamic nature of its binding to the PM (36). But the rapid kinetics of βγ shuttling and translocation here is more reminiscent of protein kinase C translocation to the PM from the cytosol in response to Ca2+ increase (t½ < 10 s) (37, 38), although reverse translocation of βγ occurs directly in response to receptor inactivation.
It is possible that an acylation cycle similar to that recently discovered in Ras (17, 26) may be at the basis of the shuttling and translocation of G protein subunits (41). YFP-tagged Hand N-Ras were shown to be present in the Golgi and in FRAP experiments to show retrograde movement from the PM to the Golgi (17, 26). In the case of G proteins, a high rate of α subunit acylation and deacylation could potentially mediate G protein shuttling. Although the known palmitoyl turnover rates of α subunits are not in agreement, the rapid FRAP kinetics of a 32-residue N-terminal peptide specific to the αi1 subunit (17) is consistent with such a likelihood. However, α subunit acylation cycling cannot explain 2BP inhibition of receptor-induced βγ translocation (41) because the α subunit never cotranslocates with βγ. Alternative possibilities are that βγ subunit acylation has remained unidentified or an unknown protein that undergoes acylation cycles binds to the βγ complex. Importantly, given the known effects of 2BP on other cellular processes (13, 27), there is also the possibility that 2BP acts on an unidentified cellular process that has an effect on G protein movement.
It is notable that lipidated hydrophobic G protein subunits are able to diffuse rapidly through the cytosol. It is possible that an unidentified protein masks the lipid moieties, allowing cytosolic diffusion. Protein partners that facilitate cytosolic trafficking by masking the hydrophobic prenyl moiety are known; for instance, specific guanine nucleotide dissociation inhibitors in the case of Rab and Rho proteins, which are prenylated similar to βγ (39, 40). Several potential candidates in the form of G protein subunit-binding proteins are known whose functions are not fully defined (11).
Overall, the shuttling of G protein subunits seen here allows heterotrimeric G proteins to continuously and rapidly test the cytoplasmic surfaces of the PM and internal membranes, maintain populations of G proteins in intracellular membranes, differentially act at a distance in response to receptor activation, and reverse on receptor inactivation, properties that appear to be uniquely suited for G protein-coupled receptor signaling and have not been anticipated previously.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health Grant GM 69027 and an American Heart Association post-doctoral fellowship (to M. C.).
The on-line version of this article (available at http://www.jbc.org) contain supplemental Figs. 1–9.
The abbreviations used are: PM, plasma membrane; FRAP, fluorescence recovery after photobleaching; FLIP, fluorescence loss in photobleaching; FRET, fluorescence resonance energy transfer; 2BP, 2-bromopalmitate; PA, photoactivation; CHO, Chinese hamster ovary; GFP, green fluorescent protein; YFP, yellow fluorescent protein; CFP, cyan fluorescent protein.
References
- 1.Gilman AG. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 2.Simon MI, Strathmann MP, Gautam N. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 3.Neves SR, Ram PT, Iyengar R. Science. 2002;296:1636–1639. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- 4.Cabrera-Vera TM, Vanhauwe J, Thomas TO, Medkova M, Preininger A, Mazzoni MR, Hamm HE. Endocr Rev. 2003;24:765–781. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- 5.Chen CA, Manning DR. Oncogene. 2001;20:1643–1652. doi: 10.1038/sj.onc.1204185. [DOI] [PubMed] [Google Scholar]
- 6.Ercolani L, Stow JL, Boyle JF, Holtzman EJ, Lin H, Grove JR, Ausiello DA. Proc Natl Acad Sci U S A. 1990;87:4635–4639. doi: 10.1073/pnas.87.12.4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leyte A, Barr FA, Kehlenbach RH, Huttner WB. EMBO J. 1992;11:4795–4804. doi: 10.1002/j.1460-2075.1992.tb05585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pimplikar SW, Simons K. Nature. 1993;362:456–458. doi: 10.1038/362456a0. [DOI] [PubMed] [Google Scholar]
- 9.Stow JL, Heimann K. Biochim Biophys Acta. 1998;1404:161–171. doi: 10.1016/s0167-4889(98)00055-x. [DOI] [PubMed] [Google Scholar]
- 10.Le-Niculescu H, Niesman I, Fischer T, DeVries L, Farquhar MG. J Biol Chem. 2005;280:22012–22020. doi: 10.1074/jbc.M501833200. [DOI] [PubMed] [Google Scholar]
- 11.Sato M, Blumer JB, Simon V, Lanier SM. Annu Rev Pharmacol Toxicol. 2006;46:151–187. doi: 10.1146/annurev.pharmtox.46.120604.141115. [DOI] [PubMed] [Google Scholar]
- 12.Ando R, Mizuno H, Miyawaki A. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 13.Resh MD. Methods. 2006;40:191–197. doi: 10.1016/j.ymeth.2006.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azpiazu I, Gautam N. J Biol Chem. 2004;279:27709–27718. doi: 10.1074/jbc.M403712200. [DOI] [PubMed] [Google Scholar]
- 15.Azpiazu I, Cruzblanca H, Li P, Linder M, Zhuo M, Gautam N. J Biol Chem. 1999;274:35305–35308. doi: 10.1074/jbc.274.50.35305. [DOI] [PubMed] [Google Scholar]
- 16.Gautam N, Downes GB, Yan K, Kisselev O. Cell Signal. 1998;10:447–455. doi: 10.1016/s0898-6568(98)00006-0. [DOI] [PubMed] [Google Scholar]
- 17.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 18.Akgoz M, Kalyanaraman V, Gautam N. Cell Signal. 2006;18:1758–1768. doi: 10.1016/j.cellsig.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirschberg K, Miller CM, Ellenberg J, Presley JF, Siggia ED, Phair RD, Lippincott-Schwartz J. J Cell Biol. 1998;143:1485–1503. doi: 10.1083/jcb.143.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang GF, Driouich A, Staehelin LA. J Cell Sci. 1993;104:819–831. doi: 10.1242/jcs.104.3.819. [DOI] [PubMed] [Google Scholar]
- 21.Choudhury A, Dominguez M, Puri V, Sharma DK, Narita K, Wheatley CL, Marks DL, Pagano RE. J Clin Investig. 2002;109:1541–1550. doi: 10.1172/JCI15420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Punnonen EL, Ryhanen K, Marjomaki VS. Eur J Cell Biol. 1998;75:344–352. doi: 10.1016/s0171-9335(98)80067-8. [DOI] [PubMed] [Google Scholar]
- 23.Chen CA, Manning DR. J Biol Chem. 2000;275:23516–23522. doi: 10.1074/jbc.M003439200. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Windh RT, Chen CA, Manning DR. J Biol Chem. 1999;274:37435–37442. doi: 10.1074/jbc.274.52.37435. [DOI] [PubMed] [Google Scholar]
- 25.Wedegaertner PB, Wilson PT, Bourne HR. J Biol Chem. 1995;270:503–506. doi: 10.1074/jbc.270.2.503. [DOI] [PubMed] [Google Scholar]
- 26.Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, Kenworthy AK. J Cell Biol. 2005;170:261–272. doi: 10.1083/jcb.200502063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vukmirica J, Tran K, Liang X, Shan J, Yuan J, Miskie BA, Hegele RA, Resh MD, Yao Z. J Biol Chem. 2003;278:14153–14161. doi: 10.1074/jbc.M211995200. [DOI] [PubMed] [Google Scholar]
- 28.Mikic I, Planey S, Zhang J, Ceballos C, Seron T, von Massenbach B, Watson R, Callaway S, McDonough PM, Price JH, Hunter E, Zacharias D. Methods Enzymol. 2006;414:150–187. doi: 10.1016/S0076-6879(06)14010-0. [DOI] [PubMed] [Google Scholar]
- 29.Michaelson D, Ahearn I, Bergo M, Young S, Philips M. Mol Biol Cell. 2002;13:3294–3302. doi: 10.1091/mbc.E02-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takida S, Wedegaertner PB. J Biol Chem. 2003;278:17284–17290. doi: 10.1074/jbc.M213239200. [DOI] [PubMed] [Google Scholar]
- 31.Quatela SE, Philips MR. Curr Opin Cell Biol. 2006;18:162–167. doi: 10.1016/j.ceb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Rocks O, Peyker A, Bastiaens PI. Curr Opin Cell Biol. 2006;18:351–357. doi: 10.1016/j.ceb.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Bivona TG, Perez De Castro I, Ahearn IM, Grana TM, Chiu VK, Lockyer PJ, Cullen PJ, Pellicer A, Cox AD, Philips MR. Nature. 2003;424:694–698. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- 34.Schroeder H, Leventis R, Rex S, Schelhaas M, Nagele E, Waldmann H, Silvius JR. Biochemistry. 1997;36:13102–13109. doi: 10.1021/bi9709497. [DOI] [PubMed] [Google Scholar]
- 35.Fivaz M, Meyer T. J Cell Biol. 2005;170:429–441. doi: 10.1083/jcb.200409157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silvius JR, Bhagatji P, Leventis R, Terrone D. Mol Biol Cell. 2006;17:192–202. doi: 10.1091/mbc.E05-05-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teruel MN, Meyer T. Cell. 2000;103:181–184. doi: 10.1016/s0092-8674(00)00109-4. [DOI] [PubMed] [Google Scholar]
- 38.Schechtman D, Craske ML, Kheifets V, Meyer T, Schechtman J, Mochly-Rosen D. J Biol Chem. 2004;279:15831–15840. doi: 10.1074/jbc.M310696200. [DOI] [PubMed] [Google Scholar]
- 39.Wu SK, Zeng K, Wilson IA, Balch WE. Trends Biochem Sci. 1996;21:472–476. doi: 10.1016/s0968-0004(96)10062-1. [DOI] [PubMed] [Google Scholar]
- 40.DerMardirossian C, Bokoch GM. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Saini DK, Kalyanaraman V, Chisari M, Gautam N. J Biol Chem. 2007;282:24099–24108. doi: 10.1074/jbc.M701191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.