Abstract
In the early Drosophila embryo, asymmetric distribution of transcription factors, established as a consequence of translational control of their maternally-derived mRNAs, initiates pattern formation1-4. For instance, translation of the uniformly distributed maternal hunchback (hb) mRNA is inhibited at the posterior to form an anterior-to-posterior protein concentration gradient along the longitudinal axis5, 6. Inhibition of hb mRNA translation requires an mRNP complex (the NRE-complex) that consists of Nanos (Nos), Pumilio (Pum) and Brain tumor (Brat) proteins, and the Nos responsive element (NRE) present in the 3’ UTR of hb mRNA7-9. The identity of the mRNA 5’ effector protein that is responsible for this translational inhibition remained elusive. Here we show that d4EHP, a cap-binding protein which represses caudal (cad) mRNA translation10, also inhibits hb mRNA translation by interacting simultaneously with the mRNA 5’ cap structure (m7GpppN, where N is any nucleotide)11 and Brat. Thus, by regulating Cad and Hb expression, d4EHP plays a key role in establishing anterior-posterior axis polarity in the Drosophila embryo.
Transcription is globally repressed in the rapidly-dividing nuclei of early Drosophila embryos, and therefore gene expression is largely regulated by translational control of maternally-provided mRNAs1. Translation is often regulated at initiation, which occurs in multiple steps starting with the recruitment of the 40S ribosomal subunit to the 5’ end of an mRNA and resulting in the correct positioning of the 80S ribosome at the initiation codon12, 13. Recognition of the cap structure by eIF4F (composed of three subunits: eIF4E, eIF4A and eIF4G) is an integral part of this process. Moreover, eIF4G interacts both with eIF4E and the poly(A)-binding protein (PABP), thus circularizing the mRNA, which in turn is believed to promote re-initiation2, 14, 15. Consistent with their importance, eIF4E and PABP have emerged as major targets of translational regulatory mechanisms mediated by such modulator proteins as 4E-BPs and Paip215-17.
Embryonic development in many metazoans requires the activity of various maternal determinants called morphogens, whose spatial and temporal expression is tightly regulated1-4. In Drosophila, local morphogen concentrations are important for the establishment of polarity and subsequent organization of both the antero-posterior and dorso-ventral axes of the embryo. A key morphogen for antero-posterior patterning is the transcription factor Hunchback (Hb); when maternal Hb is allowed to accumulate inappropriately, posterior segmentation is blocked8, 18, 19. Two modes of translational control have been proposed for the establishment of the maternal Hb gradient: translational silencing via deadenylation20 and inhibition at the initiation step in a cap-dependent manner9.
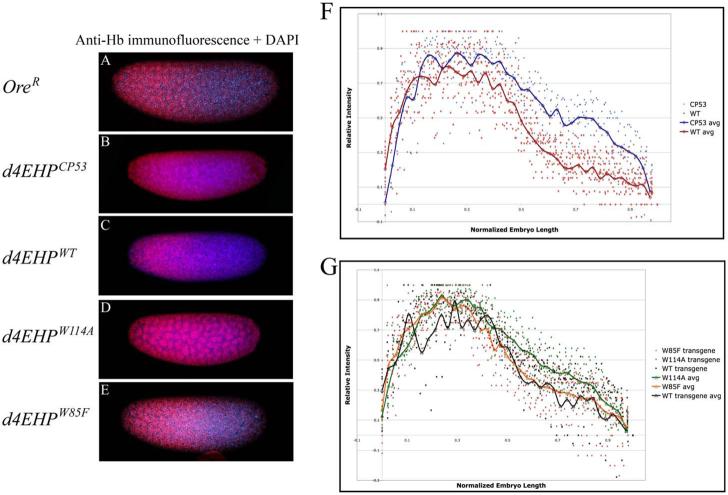
d4EHP, an eIF4E-like cap-binding protein that does not interact with deIF4G and d4E-BP, inhibits the translation of cad mRNA by interacting simultaneously with the cap and Bicoid (Bcd)10. While many embryos (∼41%) produced by females homozygous for the d4EHPCP53 mutation showed anterior patterning defects consistent with mislocalized Cad, some (∼7%) also exhibited patterning defects such as missing abdominal segments10 that cannot be readily explained by ectopic Cad expression. Since inhibition of hb mRNA translation has been linked in one study to the cap structure9 and since these additional phenotypes could be consistent with inappropriate regulation of Hb, we investigated the role of d4EHP in Hb expression. Embryos (0-2h) from females homozygous for the d4EHPCP53 mutation10 were collected and immunostained using anti-Hb antibody. DNA was stained with DAPI to highlight the nuclei (Fig. 1A-E). For simplicity, embryos will subsequently be referred to by their maternal genotype. To evaluate the extent of the Hb gradient we measured its signal intensity at 38-50 locations along the anterior-posterior axes of 6-16 embryos of each genotype. We corrected the values for overall signal intensity and then normalized the data for embryo length (EL, anterior pole = 0%, posterior pole = 100%, see Experimental Procedures). The normalized values were plotted and average intensity values were calculated to obtain an average trend (see Experimental Procedures, Fig. 1F, G). We observed that in OreR embryos, Hb signal intensity drops steeply in the middle of the embryo (Fig. 1A) and reaches 50% maximum intensity at 48% EL (Fig. 1F). In d4EHPCP53 embryos the Hb expression domain extended substantially further toward the posterior (Fig. 1B) and signal intensity remained at approximately 50% of the maximum throughout the region between 50-75% EL (Fig. 1F). Normal Hb distribution was restored to d4EHPCP53 mutant embryos by transgene-derived expression of wild-type d4EHP (d4EHPwt, Fig. 1C, G) but not by expression of a mutant form of d4EHP (d4EHPW114A), which is unable to bind the cap structure (Fig. 1D, G). Expression of another form of d4EHP (d4EHPW85F) which cannot bind Bcd, fully rescued the defective Hb gradient (Fig. 1E, G). The expression levels of the wild-type and mutant d4EHP transgenes are essentially equal10. Distributions of Nos, Pum, and Brat were unaffected in d4EHPCP53 mutant embryos (Supplemental Fig. 1). Taken together, these data demonstrate that d4EHP plays a key role in establishing the posterior boundary of Hb expression in a manner that requires its cap-binding activity but not an association with Bcd.
Fig. 1.
d4EHP interaction with the cap structure is required for hb translation inhibition. Hb immunostaining appears red, and DAPI (blue) was used to stain nuclei. A-E) Confocal images are focused on the embryo surface to illustrate the Hb gradient, and therefore not all nuclei are in focus. A) OreR embryos display normal Hb gradient. B) d4EHPCP53 mutant embryos show extended Hb expression into the posterior half. C) Expression of d4EHPWT transgene in the d4EHPCP53 mutant background rescues the mutant phenotype. D) Embryos derived from females expressing d4EHPW114A fail to fully repress posterior Hb expression. E) Embryos derived from females expressing d4EHPW85F show wild-type Hb distribution pattern. F) Plot of normalized Hb intensities measured from wt (red) and d4EHPCP53 (blue) embryos. Calculated average intensity values were used to generate average trends (thick lines). The low intensity values at the anterior pole (near 0% EL) derive from focal deformation of the image, as the focus was on the embryo surface). G) Plot of normalized Hb intensities measured from d4EHPCP53 embryos rescued with different d4EHP transgenes (d4EHPwt black, d4EHPW114A green, d4EHPW85F orange). The average trends were calculated as in F. The posterior pole is indicated as 100% (A-E) or 1 (F, G). Orientation of embryos is anterior left and dorsal up.
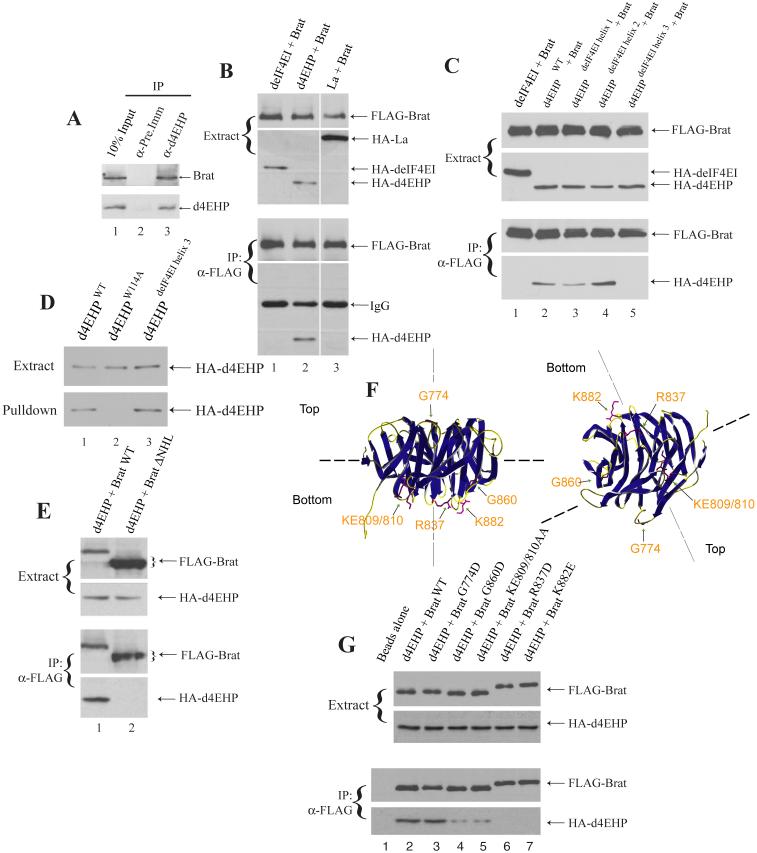
We reasoned that Brat might be a candidate partner protein for d4EHP since both are relevant for hb regulation. Thus we investigated whether d4EHP and Brat physically interact in vivo. Extracts prepared from 0-2h Oregon-R (OreR) embryos were treated with RNase and used to examine the interaction between Brat and d4EHP. Western blotting analysis using antibodies against d4EHP10 and Brat (Supplemental Fig. 2) demonstrates that, while anti-d4EHP co-immunoprecipitated endogenous Brat (Fig. 2A; lane 3), pre-immune serum did not (lane 2). To further demonstrate the specificity of this interaction, HA-tagged deIF4EI and the RNA-binding protein La (negative controls) were transfected in HEK293 cells along with FLAG-tagged full-length Brat. While anti-FLAG antibody immunoprecipitated wild-type HA-d4EHP together with FLAG-Brat (Fig. 2B, lane 2), deIF4EI and La failed to co-immunoprecipitate (lanes 1 and 3). Similarly, other RNA-binding proteins such as hnRNP U and HuR, and a d4EHP mutant (W173A), in which a tryptophan residue that is part of the hydrophobic core and thus affects protein folding is replaced, also failed to interact with Brat (data not shown), demonstrating that Brat interacts specifically with d4EHP. Since we used a cell transfection system to assay for the d4EHP:Brat interaction, it is possible that other bridging proteins are required for the d4EHP-Brat association.
Fig. 2.
Characterization of the d4EHP:Brat interaction.
A. d4EHP interacts with Brat in vivo. OreR embryo (0-2 hr) extract (lane 1) was immunoprecipitated using pre-immune (lane 2), or anti-d4EHP (lane 3) antisera. Eluted proteins were analyzed by Western blotting for the presence of Brat (top panel) and d4EHP (bottom panel). B. d4EHP interacts specifically with Brat. FLAG-tagged Brat was transfected in HEK293 cells together with HA-tagged deIF4EI, d4EHP or La. Proteins from cell extracts were immunoprecipitated (IP) with an anti-FLAG antibody and analyzed by Western blotting with antibodies against FLAG and HA. C. The d4EHP:Brat interaction is mediated by the third dorsal α-helix of d4EHP. FLAG-tagged Brat wild-type NHL domain was transfected in HEK293 cells together with HA-tagged deIF4EI, d4EHP or d4EHP/deIF4EI chimeras. B and C. Proteins from cell extracts were immunoprecipitated (IP) with an anti-FLAG antibody and analyzed by Western blotting with antibodies against FLAG and HA. D. The d4EHPdeIF4EI helix 3 mutant interacts with the cap. HEK293 cell extracts (top panel) containing transfected HA-tagged wild-type d4EHP (lane 1), d4EHPW114A (lane 2) and d4EHPdeIF4EI helix 3 (lane 3), were incubated with m7GTP-Sepharose, and the eluate was analyzed by Western blotting (bottom panel). E. d4EHP interacts with the Brat C-terminal NHL domain. FLAG-tagged Brat wild-type or ΔNHL mutant were transfected in HEK293 cells with HA-tagged d4EHP and cell extracts were subjected to Western blotting (top panel). Cell extracts were immunoprecipitated with an anti-FLAG antibody and analyzed by Western blotting (bottom panel). F. Ribbon diagrams of the Brat NHL domain21. The positions of select surface residues are indicated. G. Interaction of Brat mutants with d4EHP. FLAG-tagged wild-type (lane 2) or mutants of the Brat NHL domain (lanes 3-7) were transfected in HEK293 cells together with HA-tagged d4EHP and cell extracts were subjected to Western blotting (top panel). Cell extracts were immunoprecipitated with an anti-FLAG antibody and eluted proteins were analyzed for the presence of FLAG-Brat and HA-d4EHP by Western blotting (bottom panel). All proteins shown migrated at the positions expected from their molecular mass, as compared with molecular weight markers run on the same gels (data not shown).
To identify the Brat-interacting domain of d4EHP, we first mutated a number of individual residues located on its convex dorsal surface, and tested for co-immunoprecipitation with Brat. From this work we were unable to identify a point mutant of d4EHP that abrogated the interaction (data not shown). As an alternative approach, we created chimeric proteins in which different domains of d4EHP were replaced with their counterparts from deIF4EI, taking advantage of our knowledge that, unlike d4EHP, deIF4EI does not interact with Brat (Fig. 2C, lane 1). We produced three mutant forms of d4EHP, with each one of its three dorsal α-helices21 replaced with that of deIF4EI. We found that, while helix 1 and 2 mutants failed to disrupt binding to Brat (Fig. 2C, lanes 3 and 4), replacement of d4EHP helix 3 (residues 179 to 194) significantly reduced the interaction with Brat (Fig. 2C, lane 5). Consistent with these observations, α-helix 3 is the most divergent between d4EHP and deIF4EI10. The overall structure of d4EHP is not affected by the replacement of helix 3 with its deIF4EI counterpart, since the chimeric protein still binds to the cap (Fig. 2D, lane 3). Thus, our data demonstrate that Brat interacts with d4EHP on its convex dorsal surface and that this interaction is mediated by the third α-helix of d4EHP.
A C-terminal domain of Brat termed the NHL domain is both necessary and sufficient to inhibit hb mRNA translation7. The NHL domain contains two large surfaces (defined as top and bottom), that can support protein-protein interactions22. While the top surface of the NHL domain binds to Pum and Nos, the bottom surface does not interact with any known protein7, 22. Although the Brat NHL domain contains an amino acid sequence that conforms to the YxxxxxxLΦ d4EHP-binding motif10, the d4EHP:Brat interaction does not require this motif, since a Brat deletion mutant that lacks it can still interact with both d4EHP and the d4EHP W85F mutant (Supplemental Fig. 3). This sequence is most probably masked from interaction with d4EHP because it is located in the hydrophobic core of the NHL domain22. To determine whether the d4EHP:Brat interaction requires the NHL domain, a Brat mutant that lacks the domain (Brat ΔNHL) was engineered and used in a co-immunoprecipitation experiment (Fig. 2E). While wild-type Brat was readily co-immunoprecipitated with d4EHP, the Brat ΔNHL mutant was not (compare lanes 1 and 2). Thus, we conclude that the NHL domain is the site of d4EHP interaction. To further characterize this interaction, point mutations were designed to replace residues on the two surfaces of the NHL domain (Fig. 2F), and the mutant proteins were tested for their ability to interact with d4EHP. Mutation of a top surface residue that affects Brat interaction with Pum (G774A; Fig. 2G, lane 3)7 did not affect the d4EHP:Brat interaction. However, when residues on the bottom surface were mutated, the d4EHP:Brat interaction was either significantly reduced (G860D and KE809/810AA; lanes 4 and 5), or abrogated (R837D and K882E; lanes 6 and 7; note that the charge differences caused R837D and K882E mutant proteins to migrate slower in the gel). Importantly, the Brat NHL R837D mutant can assemble into an NRE-complex (see below; Fig. 3, lane 4), demonstrating that this mutation specifically affects the d4EHP interaction and not the interactions with Pum and Nos.
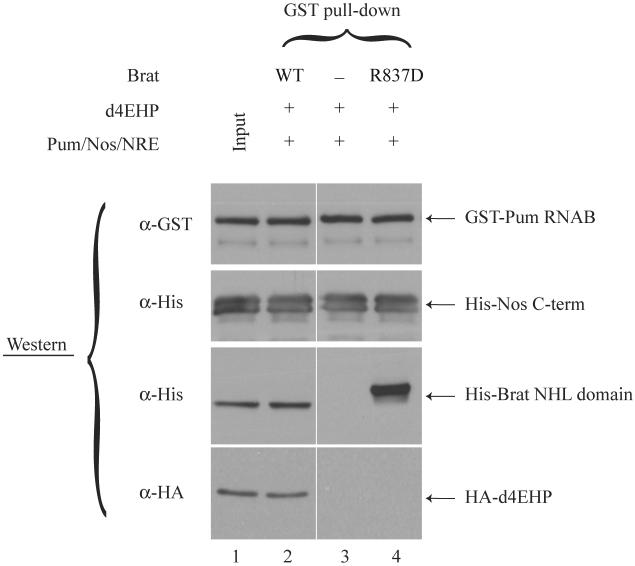
Fig. 3.
d4EHP interacts with the NRE-complex in vitro.
Samples containing in vitro translated HA-tagged d4EHP and purified components of the NRE-complex were used to perform an in vitro GST pull-down experiment. Eluted proteins were analyzed for the presence of GST-Pum RNAB, His6-Nos C-term, His6-Brat NHL domain and HA-d4EHP by Western blotting. All proteins shown migrated at the positions expected from their molecular mass, as compared with molecular weight markers run on the same gels (data not shown).
Brat inhibits hb mRNA translation by interacting with the NRE-complex7. Since d4EHP interacts physically with Brat, we asked whether d4EHP can be co-purified with the NRE complex in vitro. Incubation of recombinant components of the NRE-complex (Brat, Pum, Nos and NRE) together with HA-tagged d4EHP resulted in the retention of d4EHP on glutathione-Sepharose beads through the GST-Pum RNAB fusion protein (Fig. 3, lane 2). The association of Brat with d4EHP was dependent on the ability of d4EHP to bind to Brat, since addition of Pum/Nos/NRE alone or in combination with the Brat R837D mutant failed to capture it (lanes 3 and 4). Thus, by interacting with Brat, d4EHP can associate with the NRE complex.
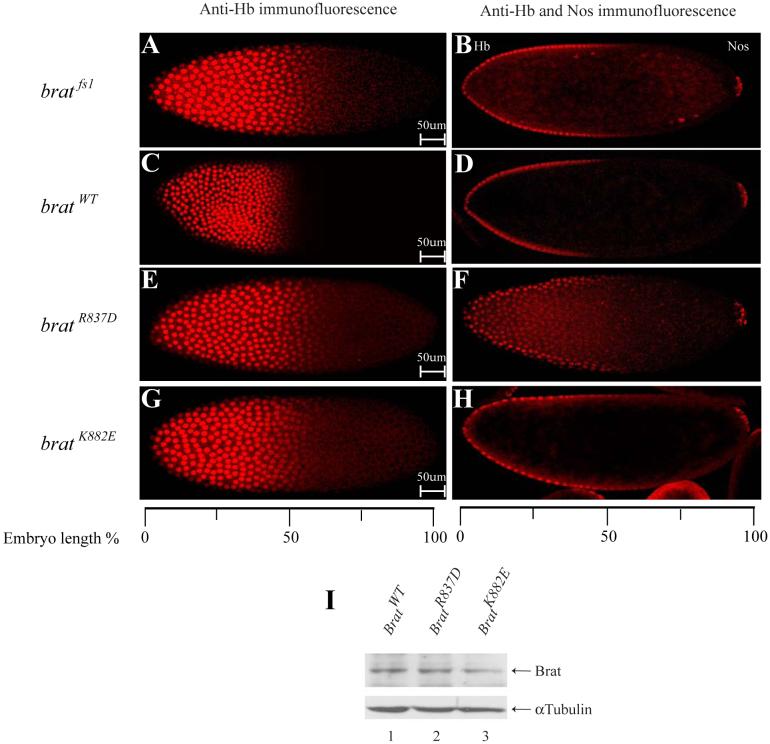
To investigate the biological significance of the d4EHP:Brat interaction, we studied the effects of Brat mutants, which are defective for d4EHP binding, in Drosophila embryos. As previously shown7, bratfs1 mutant embryos exhibit a significant expansion of the Hb expression domain towards the posterior (Fig. 4A and 4B) and display severe abdominal segmentation defects (Table 1). When a bratWT transgene is expressed in the bratfs1 mutant background, normal Hb distribution (Fig. 4C and 4D) and a wild-type segmentation pattern is restored7 (Table 1). To investigate whether interaction with d4EHP is essential for the function of Brat in embryonic patterning, we introduced transgenes encoding mutant forms of Brat that affect the d4EHP:Brat interaction (bratR837D and bratK882E) into the bratfs1 mutant background. Despite being expressed at levels similar to the bratWT transgene (Fig. 4I), these mutant forms fail to fully rescue the normal Hb gradient (Fig. 4E-4H) and, importantly, do not fully rescue the bratfs1 mutant phenotype (Table 1). Taken together, our data strongly argue that the d4EHP:Brat interaction contributes significantly to hb regulation.
Fig. 4.
Functional analysis of brat mutants in transgenic Drosophila embryos.
A and B. Embryos derived from homozygous bratfs1 females show a shift of the Hb expression boundary towards the posterior. C and D. Embryos derived from females expressing bratWT in the bratfs1 mutant background show wild-type Hb distribution pattern. E-H. Embryos derived from females expressing mutant bratR837D and bratK882E genes (unable to bind d4EHP) exhibit ectopic Hb expression. B, D, F and H. Embryos stained for both Hb and Nos proteins using the same secondary antibody, the latter serving as an internal control for staining intensity. The anterior tip is indicated as 0%. Orientation of embryos is anterior left and dorsal up. I. Western blot analysis of embryo extracts using anti-Brat or anti-α-tubulin as a loading control. Two to three independent transgenic lines were examined for each experiment, with similar results.
Table 3.1.
Abdominal segmentation defects in Brat mutant embryos
| No. of abdominal segments |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| Bratfs1/Df(2L)TE37C-7 | 2 | 10 | 62 | 20 | 5 | 1 | |||
| Bratfs1/Df(2L)TE37C-7; BratWT | 100 | ||||||||
| Bratfs1/Df(2L)TE37C-7; BratR837D | 7 | 24 | 39 | 30 | |||||
| Bratfs1/Df(2L)TE37C-7; BratK882E | 20 | 41 | 39 | ||||||
Each entry is the percentage of embryos derived from females of the indicated genotype (left) bearing the indicated number of abdominal segments (above). Seventy to one-hundred embryos were scored in each case.
We have demonstrated here that through its interaction with Brat, d4EHP defines and sharpens the posterior boundary of Hb expression. Based on the hypomorphic d4EHPCP53 phenotype, its activity appears most relevant to hb regulation in the region of the embryo from 50-75% EL, although it is possible that a null d4EHP allele would have more drastic effects. The d4EHP:Brat interaction is mediated via residues on the bottom surface of the Brat NHL domain (Fig. 2F,G). Thus, as in the model we established for cad10, a simultaneous interaction of d4EHP with the cap and Brat results in mRNA circularization and renders hb translationally inactive. Since the interaction between Brat and d4EHP does not involve the 4EHP-binding motif we previously described (YxxxxxxLΦ), it is possible that d4EHP interacts with Brat through a bridging protein.
Our data support a model for the requirement for the 5’ cap structure in regulation of endogenous hb mRNA. This is consistent with an earlier study that assessed translation of NRE-containing mRNAs after injection into Drosophila embryos and concluded that the cap structure is functionally significant9. In contrast, another study reported that Nos and Pum repressed the expression of an engineered transgene containing an internal ribosome entry site (IRES) and a hairpin loop designed to block cap-dependent translation23. These results were used to conclude that hb translational repression is cap-independent. However, the phenotypic assay used in that study was indirect and the observed results could also be caused by RNA destabilization. Furthermore, Nos-dependent deadenylation was also shown to be important in establishing the Hb gradient20. It is difficult to reconcile all these data without concluding that multiple distinct post-transcriptional mechanisms regulate Hb expression, including two that require Nos. The novel d4EHP-dependent mechanism we defined appears important for repressing hb in more central regions of the embryo, while cap-independent regulation involving deadenylation of hb mRNA may predominate in more posterior regions of the embryo. We note that mutant forms of Brat that are abrogated for d4EHP interaction retain substantial (but not complete) activity in repressing hb, suggesting some redundancy between these two mechanisms. Analogous overlapping translational control mechanisms have recently been reported for Bruno, which represses Oskar (Osk) expression both through cap-dependent translational regulation and through packaging osk mRNA into translationally silent RNP complexes24.
Our identification of a common inhibitory mechanism which regulates cad and hb mRNA translation simplifies our understanding of how the anterior-posterior axis is organized during early Drosophila embryogenesis. By regulating two classical maternal morphogenetic gradients, d4EHP plays a critical role in early Drosophila embryonic development. It is noteworthy that d4EHP is recruited to these mRNAs through different RNA binding proteins that presumably recognize different sequence elements. In the case of cad, d4EHP becomes associated by binding directly to Bcd, which in turn recognizes a defined 3’UTR element, the BBR25, 26. In the case of hb, Bcd binding is not involved in d4EHP recruitment and no element similar to the BBR is present. It remains uncertain whether the interaction between d4EHP and Brat is direct or indirect; as d4EHP and Brat are both uniformly distributed in early embryos7, 10, a non-uniformly distributed bridging protein mediating this interaction may be the basis of the spatially-restricted requirement for d4EHP in hb repression. Since d4EHP and some of its interacting partners are evolutionarily conserved in higher eukaryotes and because cap-dependent translation regulation plays such an important role in eukaryotic gene expression16, we predict that 4EHP-dependent translational inhibitory mechanisms are widespread throughout the animal kingdom.
Experimental Procedures
Plasmids
Cloning of d4EHP was previously described10. Brat cDNA (RE16276; Research Genetics) was obtained from the Berkeley Drosophila Genome Project27. All constructs reported herein were produced using the polymerase chain reaction (PCR). For brat, PCR-amplified wild-type and mutant cDNAs were introduced into the pcDNA3-N-term-FLAG vector using EcoRV/NotI sites. For recombinant protein expression, Brat NHL domain and Nos C-term domain (Nos C-term) were subcloned into the pProEx-His vector using SalI/NotI and EcoRI/XhoI sites, respectively, and Pum RNA-binding domain (Pum RNAB) into the pGEX 6p-1 vector using EcoRI/SalI sites. NRE from hb mRNA, flanked by XbaI sites, was introduced into the 3’UTR of pcDNA3-rLuc-ΔApaI reporter vector. To create pCaSpeR4-nos promoter-Brat wild-type and mutant rescue vectors, Brat constructs were inserted into the pKS-nos promoter vector using NheI/NotI sites. Subsequently, a Kpn1/NotI cassette from pKS-nos promoter-Brat wild-type and mutant vectors were transferred into the pCaSpeR4 vector. All inserts were fully sequenced.
Recombinant Protein Purification
E. coli BL21(DE3) transformed with the pProEx-Brat NHL domain, pProEx-Nos C-term and pGEX-Pum RNAB constructs were used to produce His-Brat NHL domain, His-Nos C-term and GST-Pum RNAB fusion proteins as previously described10. TALON™ Metal Affinity resin (BD Bioscience) and Glutathione Sepharose™ 4B resin (Amersham Pharmacia) were used according to the manufacturer’s instructions.
Anti-Brat antibody and Western Blotting Analysis
An anti-Brat antibody (#3187) was raised in a New Zealand White rabbit injected with recombinant His-Brat NHL domain protein and used for Western blotting (1:3000). Cell culture, Co-immunoprecipitation and Western blotting were performed as previously described10.
Transgenic Rescue Experiment
Transgenic flies were generated by P-element mediated germline transformation of yw recipients using pCaSpeR-nos promoter-Brat wild-type and mutant rescue vectors. Transformed brat lines were crossed to the bratfs1 mutant and tested for the rescue of mutant phenotypes. pUASp-d4EHP transgenic lines10 and antibody staining were performed as previously described28. Hb, and Nos, Pum and Brat immunostainings were visualized using AlexaFluor® 546 goat anti-rat IgG secondary and AlexaFluor® 488 goat anti-rabbit IgG secondary, respectively (1:500; Molecular Probes) using a confocal laser scanning microscope. Embryo images were analyzed for Hb gradient using Zeiss LSM data acquisition software.
In Vitro Transcription/Translation and Binding Assay
pcDNA3-3HA-d4EHP10 and pcDNA3-rLuc-ΔApaI-NRE vectors were linearized with ApaI and transcribed using T7 RNA polymerase (MBI). Nuclease treated rabbit reticulocyte lysate (Promega) was incubated for 1h at 30°C with 300ng of HA-d4EHP mRNA. Subsequently, the extract was supplemented with components of the NRE-complex and the experiments of Fig. 4 were performed as previously described7, 29.
Image Analyses, Immunofluorescence Quantitation, and Data Analyses
0-2 hrs old embryos were immunostained with anti-Hb (1:10,000, a gift of P. Macdonald) and AlexaFluor® 546 conjugated secondary antibodies (Molecular Probes) and DNA was stained with DAPI. Images were captured using a Zeiss LSM510 confocal microscope. Quantitation of the fluorescence intensity was performed with Openlab™ (Improvision) by recording the intensity values within a nucleus-sized area sliding along the anterior-posterior axis. Multiple embryos (6-16) for each genotype were measured at 38-50 positions along the anterior-posterior axis for a total of more than 3000 data points. For each embryo, length was normalized (0%= anterior pole, 100%= posterior pole) and measured intensities were normalized by subtracting the intensity recorded at the posterior pole and dividing this value by the maximum intensity measured within the same embryo. Individual data points could have values below zero if the local signal intensity was less than that at the posterior pole. To generate the average curve, the data points for each genotype were grouped in 38 bins (corresponding to the smallest sample set). For each bin, values were averaged and the resulting 38 points constitute the average curve.
Supplementary Material
Supplemental Fig. 1: Distribution pattern of the components of the NRE-complex. Nos, Pum and Brat distribution in OreR (A-C) and d4EHPCP53 mutant (D-F) embryos were visualized via immunofluorescence. Antibodies were used at 1:500 dilutions. Orientation of embryos is anterior left and dorsal up.
Supplemental Fig. 2: Characterization of anti-Brat antibody.
A. Brat antiserum detects the recombinant Brat NHL domain. Recombinant His-tagged Brat NHL domain (lane 1) is detected in a Western blot with a Brat antiserum (lane 3), but not with pre-immune serum (lane 2). B. Brat antiserum immunoprecipitates Brat from cell extracts. FLAG-tagged Brat was transfected in HEK293 cells (lane 1) and immunoprecipitated with an anti-FLAG antibody (lane 2), pre-immune serum (lane 3), or Brat antiserum (lane 4).
Supplemental Fig. 3: The d4EHP:Brat interaction does not require the YxxxxxxL Φ d4EHP-binding motif. FLAG-tagged Brat wild-type or ΔBind mutant were transfected in HEK293 cells with HA-tagged deIF4EI, d4EHP wild-type and d4EHP W85F mutant and cell extracts were subjected to Western blotting (Total extract). Cell extracts were immunoprecipitated (IP) with an anti-FLAG antibody and analyzed by Western blot (IP panels).
Acknowledgements
We thank P. Macdonald (anti-Hb), N. Dostatny (pKS-nos-X-Bcd 3’UTR vector) and Z. Lev (bratfs1) for reagents; J. Laliberte for confocal microscopy; Guillaume Lesage in the Cell Imaging and Analysis Network (CIAN) and Jelena Jevtic for valuable help with the image analyses and quantification; Y. R. Lee, H. Beili, B. Spelke, and C. Lister for technical assistance; F. Poulin, M. Bidinosti, J. Chicoine, M. Ferraiuolo, B. Raught, A-C. Gingras, L. Huck, K. Madjar and other members of our labs for valuable discussions. Y.A.C-P. was supported by the Canadian Institute of Health Research (CIHR) Burroughs Wellcome scholarship to medical students. N.S. is a CIHR Distinguished Scientist and a Howard Hughes Medical Institute (HHMI) International Scholar. This work was supported by grants from the CIHR to N.S. and P.L, by a grant from the NCIC with funds from the Canadian Cancer Society to P. L., and by a grant from the NIH (R01 GM66157) to N.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wickens M, Goodwin EB, Kimble J, Strickland S, Hentze M. Translational Control of Gene Expression. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 295–370. [Google Scholar]
- 2.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–35. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.St Johnston D, Nusslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992;68:201–19. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- 4.Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- 5.Tautz D, et al. Finger protein of novel structure encoded by hunchback, a second member of the gap class of Drosophila segmentation genes. Nature. 1987;327:383–89. [Google Scholar]
- 6.Tautz D. Regulation of the Drosophila segmentation gene hunchback by two maternal morphogenetic centres. Nature. 1988;332:281–4. doi: 10.1038/332281a0. [DOI] [PubMed] [Google Scholar]
- 7.Sonoda J, Wharton RP. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15:762–73. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wharton RP, Struhl G. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell. 1991;67:955–67. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- 9.Chagnovich D, Lehmann R. Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc Natl Acad Sci U S A. 2001;98:11359–64. doi: 10.1073/pnas.201284398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho PF, et al. A new paradigm for translational control: inhibition via 5’-3’ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005;121:411–23. doi: 10.1016/j.cell.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 11.Shatkin AJ. Capping of eukaryotic mRNAs. Cell. 1976;9:645–53. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 12.Hershey JWB, Merrick WC. In: Translational Control of Gene Expression. Sonenberg N, Hershey JWB, Mathews MB, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 33–88. [Google Scholar]
- 13.Poulin F, Sonenberg N. In: Translation Mechanisms. Lapointe J, Brakier-Gingras L, editors. Landes Bioscience; Austin, Texas: 2003. pp. 280–297. [Google Scholar]
- 14.Sachs A. In: Translational Control of Gene Expression. Sonenberg N, Hershey JWB, Mathews MB, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 447–466. [Google Scholar]
- 15.Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou M,N, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–13. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–80. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 17.Khaleghpour K, et al. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell. 2001;7:205–16. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 18.Struhl G, Johnston P, Lawrence PA. Control of Drosophila body pattern by the hunchback morphogen gradient. Cell. 1992;69:237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann R, Nusslein-Volhard C. hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev Biol. 1987;119:402–17. doi: 10.1016/0012-1606(87)90045-5. [DOI] [PubMed] [Google Scholar]
- 20.Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, Strickland S. Nanos and Pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124:3015–23. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- 21.Marcotrigiano J, Gingras A-C, Sonenberg N, Burley SK. Cocrystal structure of the messenger RNA 5’ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–61. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 22.Edwards TA, Wilkinson BD, Wharton RP, Aggarwal AK. Model of the Brain Tumor-Pumilio translation repressor complex. Genes Dev. 2003;17:2508–13. doi: 10.1101/gad.1119403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wharton RP, Sonoda J, Lee T, Patterson M, Murata Y. The Pumilio RNA-binding domain is also a translational regulator. Mol Cell. 1998;1:863–72. doi: 10.1016/s1097-2765(00)80085-4. [DOI] [PubMed] [Google Scholar]
- 24.Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–33. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 25.Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379:694–9. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 26.Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring WJ, Jäckle H. RNA binding and translational suppression by Bicoid. Nature. 1996;379:746–9. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 27.Rubin GM, et al. A Drosophila complementary DNA resource. Science. 2000;287:2222–4. doi: 10.1126/science.287.5461.2222. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi S, Amikura R, Nakamura A, Lasko PF. In: Advances in Molecular Biology: A Comparative Methods Approach to the Study of Oocytes and Embryos. Richter J, editor. Oxford University Press; NY: 1999. pp. 426–445. [Google Scholar]
- 29.Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–12. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1: Distribution pattern of the components of the NRE-complex. Nos, Pum and Brat distribution in OreR (A-C) and d4EHPCP53 mutant (D-F) embryos were visualized via immunofluorescence. Antibodies were used at 1:500 dilutions. Orientation of embryos is anterior left and dorsal up.
Supplemental Fig. 2: Characterization of anti-Brat antibody.
A. Brat antiserum detects the recombinant Brat NHL domain. Recombinant His-tagged Brat NHL domain (lane 1) is detected in a Western blot with a Brat antiserum (lane 3), but not with pre-immune serum (lane 2). B. Brat antiserum immunoprecipitates Brat from cell extracts. FLAG-tagged Brat was transfected in HEK293 cells (lane 1) and immunoprecipitated with an anti-FLAG antibody (lane 2), pre-immune serum (lane 3), or Brat antiserum (lane 4).
Supplemental Fig. 3: The d4EHP:Brat interaction does not require the YxxxxxxL Φ d4EHP-binding motif. FLAG-tagged Brat wild-type or ΔBind mutant were transfected in HEK293 cells with HA-tagged deIF4EI, d4EHP wild-type and d4EHP W85F mutant and cell extracts were subjected to Western blotting (Total extract). Cell extracts were immunoprecipitated (IP) with an anti-FLAG antibody and analyzed by Western blot (IP panels).