Abstract
The Ensembl project (http://www.ensembl.org) is a comprehensive genome information system featuring an integrated set of genome annotation, databases and other information for chordate and selected model organism and disease vector genomes. As of release 47 (October 2007), Ensembl fully supports 35 species, with preliminary support for six additional species. New species in the past year include platypus and horse. Major additions and improvements to Ensembl since our previous report include extensive support for functional genomics data in the form of a specialized functional genomics database, genome-wide maps of protein–DNA interactions and the Ensembl regulatory build; support for customization of the Ensembl web interface through the addition of user accounts and user groups; and increased support for genome resequencing. We have also introduced new comparative genomics-based data mining options and report on the continued development of our software infrastructure.
INTRODUCTION
The availability of complete genome sequences for an increasing number of chordates has had a dramatic impact on biomedical research in the 21st century. Now 7 years beyond the initial publications of the draft human genome sequence (1,2), both the number of sequenced genomes and the total amount of genome-wide data that can be naturally organized on the genome sequence continue to rapidly increase. The Ensembl project provides a comprehensive genome information system consisting of data storage, integration, analysis and visualization of a wide variety of biological data. In comparison to similar projects based at the University of California Santa Cruz (3) and the National Center for Biotechnology Information (4) the distinguishing characteristics of the Ensembl project include:
– The Ensembl genome browser available at http://www.ensembl.org providing visualization for our own and collaborators genome annotations, alignments, variation and functional genomics data and supporting additional data integration through the DAS protocol.
– Ensembl gene sets created using an automated analysis pipeline that has been significantly optimized based on the completeness of the genome sequence and the availability of species-specific supporting data.
– The Ensembl application programming interface (API) that allows programmatic access to all of our data sets including annotations, genomic alignments and variation data.
– BioMart data mining tools, which support sophisticated Ensembl-specific queries and federated queries with other BioMart-compliant data resources.
– An entirely open resource with all of our code and data freely available to all users.
Ensembl data is organized into several species-specific and multi-species MySQL databases. Each database is named using the format <species>_<database type>_<release number>_<data version>. For each supported species, a core database contains the DNA sequences, gene annotations, external references, etc. Databases of type ‘otherfeatures’ are provided for each supported species (except for the low-coverage genomes) and include EST genes, external annotation sets and other data. Variation databases that include dbSNP (5) and resequencing data (see subsequently), are provided for 10 species. This year, we introduced a functional genomics database, initially released for human and mouse, to support functional data types assayed by whole-genome tiling arrays or high-throughput sequencing (see subsequently). Comparative genomics data and the supporting data for the Ensembl BioMart datamining tool (6) are provided in multi-species databases.
Ensembl generally releases updates six times each year in February, April, June, August, October and December. Specific data updates are driven by the availability of new or updated genome sequence assemblies, significant increases in supporting evidence for genome annotations, updated releases of major external data sets [such as dbSNP (5)] that are incorporated into Ensembl, and new biological data resources such as protein–DNA interaction maps based on genome-wide ChIP-chip and ChIP-seq data sets. Each new Ensembl release may also include new data visualization options and improvements to the underlying software infrastructure.
This report lists only some of the new features, new data and other improvements that we have added to Ensembl since our last report (7). Users interested in the most up-to-date details of the Ensembl project should visit the Ensembl main page (http://www.ensembl.org) and follow the ‘What's new’ link and/or subscribe to the low-volume ‘Ensembl announce’ mailing list by sending email ‘subscribe ensembl-announce’ as the message body to majordomo@ebi.ac.uk. Other information about Ensembl features is available on the Ensembl help pages or by email at helpdesk@ensembl.org.
RESULTS
Ensembl regulatory build
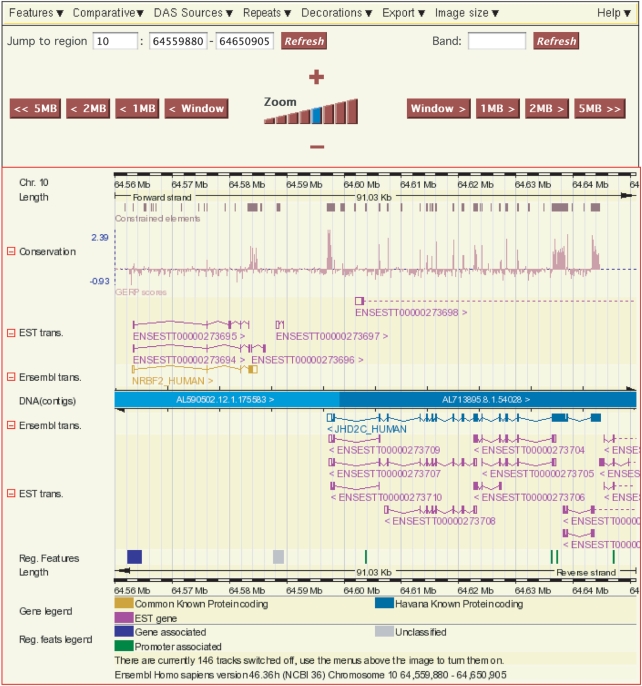
The Ensembl regulatory build is designed to automatically annotate all of the functional regulatory regions in the genome and assign putative functions to as many of these regions as possible. The initial release of the Ensembl regulatory build in June 2007, integrated eight genome-wide data sets, mainly in pre-publication ‘resource’ status, to identify ∼110 000 regulatory features across the human genome. Briefly, the integration procedure starts with likely regulatory regions (such as DNase I hypersensitive sites) and seeks to identify the function of each site by analysing specific patterns of histone modification immediately adjacent to the region. We identified a number of patterns highly enriched for gene starts, genic regions and distal regions. Ensembl regulatory features are displayed on ContigView (Figure 1).
Figure 1.
The Ensembl regulatory build and GERP conservation track. A 90 kb region of human chromosome 10 showing Ensembl regulatory features in blue, green and grey on the bottom track and the GERP conservation track at the top. Note the overlap of gene-associated regulatory features with the start regions of both Ensembl transcripts and EST transcripts suggesting a complex transcriptional environment. The conservation track is a composite track that displays the constrained elements by default and both constrained elements and the GERP scores when expanded.
Functional genomics database
As noted above, the Ensembl Functional Genomics Database is the fourth species-specific database that is part of the standard Ensembl release. The Functional Genomics Database and its associated API provide a platform for the storage, analysis and visualization of array-based functional genomics data. We have created an initial infrastructure for analysis of these data based on the Ensembl analysis pipeline (8). This structure supports the modular incorporation of analysis tools dedicated to various aspects of tiling array analysis such as normalization and platform-specific hit identification.
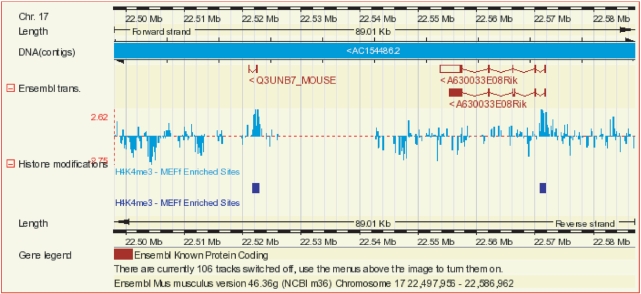
The database is currently used to support the Ensembl regulatory build (see above) and the display on of ChIP-chip data and analysis within Ensembl (Figure 2). The database and API feature a fully automated data import structure, an extensible array model and support for the Tab2MAGE metadata format (9). Additionally, the database is designed for deployment in external research laboratories and supports local data processing and visualization through DAS.
Figure 2.
ChIP-chip display. Histone 3 lysine trimethylation data from mouse embryonic fibroblast cells (29) on mouse chromosome 17 in the region of the Q3UNB7 (ENSMUSG00000073442) and A630033E08Rik (ENSMUSG00000059142) genes. The Histone modifications display is a composite track that combines raw enrichment values and peak identifications. Displays that encompass large regions of the genome include only the identified peak regions.
Ensembl customization: user accounts and groups
The major new Ensembl website functionality over the past year is the addition of user and group accounts. These accounts enable users to create bookmarks, customize their Ensembl interface and share their bookmarks and configurations with other users in an Ensembl group. We note, importantly, that all Ensembl data is equally accessible to users whether or not they create an user account.
Ensembl user accounts are designed to personalize the Ensembl interface. As the number of data tracks in Ensembl has grown, the default visualization settings are not ideal for every user. For example, some users may be interested in displaying only the Ensembl genes track together with mapping of gene expression arrays and SNP locations, while other users may want a display consisting of constrained elements, RNA genes, the underlying clone tilepath, or any of more than one hundred available data tracks. These personalized interfaces can now be saved and shared through Ensembl accounts.
Ensembl Groups have several functions. The primary function is to share configurations, bookmarks, or notes with other members of the group. Single users can also create groups as virtual folders to organize bookmarks, configurations and notes-based separate projects. Groups may be created and administered by any user with an Ensembl account. Group administrators can invite anyone to join their group and users can be members of several groups simultaneously. All group members must also have Ensembl accounts.
Notes are currently supported on GeneView pages and allow users the option of creating their own annotations and have these integrated into the web display. Notes will be added to other pages in the future.
New species and improved gene annotations
The Ensembl website currently displays data for 41 species. In the past year, we have added data for seven new high coverage genomes and generated updated gene sets for eight species. Previously, we reported that four low-coverage (2×) genome gene sets were available with five more underway (7). During this year we have finished the both the gene sets in progress and sets for an additional five species [Spermophilus tridecemlineatus (squirrel), Tupaia belangeri (tree shrew), Cavia porcellus (guinea pig), Microcebus murinus (mouse lemur), Ochotona princeps (pika)]. This set of 14 low-coverage annotated genome sequences provides an extensive resource for mammalian comparative genomics.
We have continued the CCDS (Consensus Coding Sequence) collaboration with the Sanger Institute's Havana group (http://www.sanger.ac.uk/HGP/havana/), UCSC (3) and NCBI (4). CCDS is a stable set of protein-coding gene structures for which all consortium members agree on to the base pair. We have released an update to the set that includes 18 290 CDSs from 16 003 genes. This is a substantial improvement in gene coverage over the previous set which contained 14 795 CDSs from 13 142 genes. A CCDS set has also been generated for mouse, which includes 13 374 CDSs from 13 014 genes. Further updates to CCDS sets are in progress based on new human and mouse Ensembl gene builds, Refseq (10) builds and Havana annotation. Additional details regarding the CCDS project are available from http://www.ncbi.nlm.nih.gov/CCDS/.
The Ensembl gene build process is based on alignments of protein and cDNA sequences and in order to produce a high-quality gene set, it is crucial to maximize the value of species-specific sequence data and ensure the suitability of all input sequences. In light of this, we have made improvements to several stages of the automatic annotation process. Improved use of species-specific sequences primarily addressed gene models characterized by a short first CDS exon followed by a long (>10 000 bp) intron as well as those with non GT–AG splice sites. Using standard gene-wise (11) parameters, neither case was predicted well by the Ensembl pipeline. To address these cases, we now run gene-wise with two different parameter sets and also run exonerate (12), a faster alignment algorithm more suited to the longer genomic sequences required for accurate long intron prediction. The results of these three analyses for each protein are compared and the best gene prediction chosen on the basis of a set of rules including percentage identity of the model to the original protein. Using this improved method, the percentage of Refseq genes for which we produce at least one identical CDS model increased from 78% to 88% and for Havana genes from 79% to 88%. We have also improved the quality of the input sequence data by a careful filtering process that identifies anomalous sequences such as chimeric cDNAs, cDNAs with retained introns and viral proteins, and protein sequences derived from repeats. For example, we remove from our input sequence data all of the cDNAs annotated as chimeric by the Mammalian Gene Collection (13). Removing these protein and cDNA sequences from the Ensembl gene build input reduced artefactual gene merging and over prediction.
Two other notable gene build improvements represent incorporation of information not previously used by Ensembl. The first development concerns UTRs that are added from cDNAs, when the cDNA exon boundaries match those from the protein model. Often there is a choice of possible cDNAs with differing UTRs. We are now prioritizing these cDNA choices on whether they match the boundaries of paired end tags (ditags) experimentally derived from the starts and ends of cDNAs, providing a second source of evidence to accurately determine UTR boundaries. We are mapping ditag sequences from the Genome Institute of Singapore and from the Fantom project for human and mouse (14–16). The second enhancement is specific to immunoglobulin segments, which present problems for standard gene prediction methods because the somatic rearrangements of gene segment clusters make complete cDNAs difficult to align. We now align annotated segments from the IMGT database (17) for mouse and human. The predictions based on these replace any overlapping gene models produced by the standard Ensembl pipeline in the immunoglobulin gene clusters.
New gene builds in 2007 included updates to both human and mouse, which both benefited from the methodological improvements described above. For the case of mouse, the new gene build was in support of the newly released NCBI build 37 genome assembly, while the updated human gene build incorporates the latest Havana manual annotation set.
Resequencing data: new resources and visualization
New sequencing technologies are expected to make whole genome resequencing feasible on a large scale (18,19). The genome sequence for a single individual is already available using previous generation sequencing technology (20). We recently reported on TranscriptSNPView, a transcript-based visualization for resequencing data and our SSAHA-based (21) alignment of resequencing reads to the mouse genome (22). We have extended this technique and TranscriptSNPView over the past year to include resequenced human individuals and rat strains. This year we have developed additional resources for analysis and visualization of resequencing data. The new SequenceAlignView (Figure 3) displays the reference genome sequence together with the genome sequence of individuals (or strains in the case of mouse and rat). With this view, the exact sequence of the individual can be quickly determined and the differences between the sequenced individual and the reference genome assembly highlighted. Resequencing data is also provided in structured EMF (Ensembl Multi-Format) text files. On our FTP site, users doing comparative genomics will also find EMF files available for multiple sequence alignments.
Figure 3.
SequenceAlignView. A full screen shot of a region on mouse chromosome 8 displaying available resequencing data from the 129S1/SvImJ, 129X1/SvJ and A/J laboratory mouse strains (the 129S1/SvImJ stain is marked as having no data in the region). Numerous display options are in the top panel on the page, which allow user to choose any region of the genomes, highlight Ensembl annotations, locations of knows SNPs and other information. The resequencing alignment in the bottom panel identifies exons in red and SNPs in yellow. Links to individual variations are provided to the right of the resequencing alignment.
DAS extensions
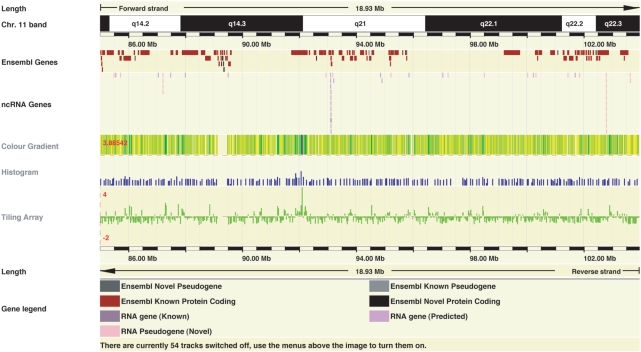
Ensembl continues to make extensive use of the DAS protocol (23). During this year, we have released two new DAS resources. Previously, we extended the Ensembl genome browser with DAS client functionality, which allows researchers around the world to remotely host data sources and view these on major Ensembl displays including CytoView, ContigView, GeneView and ProtView (24). This year, we extended our client visualization support through DAS to include a colour gradient, histogram and tiling array ‘wiggle’ format (Figure 4). These new visualization options are particularly applicable to dense genome data such as that produced by whole-genome tiling array experiments. We now also serve current Ensembl data for integration into other DAS clients. Data available for integration into our DAS clients includes transcripts, ditag data, markers, karyotype information, repeats and DNA and protein align features including cDNA alignments and UniProt alignments. DAS sources setup by Ensembl are also automatically registered with the DAS registry (25). Instructions for using DAS with Ensembl are available from http://www.ensembl.org/info/data/external_data/das/index.html.
Figure 4.
DAS Visualizations. A 19 Mb region of human chromosome 11 showing identical data displayed with (from top to bottom) the colour gradient, histogram and tiling array ‘wiggle’ format. The colour gradient format transitions from yellow (low values) to blue (high value). The histogram display format supports merged data in bins across the genome; the display value is selectable to be either the average of the bin (shown here) or the maximum value in the bin to achieve greater data contrast. In the histogram format, the lowest value in the data set becomes the baseline. The tiling array format allows for the display of both positive and negative values with overlapping data points resulting in the maximum data point being displayed. All three display formats support in-line data normalization. The ideal format will depend on the data to be displayed. These example data are P-values from a genome-wide association study (30).
Ensembl software infrastructure
The Ensembl core software system (26) provides an efficient way of representing genome data in a relational database and providing access to it via an object-oriented API. This API is used by our computational pipelines to generate and store genome annotation, and by the Ensembl website to retrieve information that is to be displayed to the user. Bioinformaticians can use the API to access Ensembl databases remotely (Ensembl databases are available at mysql://ensembldb.ensembl.org:3306; Ensembl BioMart databases use mysql://martdb.ensembl.org:3316) or local databases containing their own data. We maintain full unit test coverage for the API.
The database representation and API are being continuously developed to address bottlenecks affecting website and pipeline performance and increase flexibility. While most of this development is incremental in nature, two significant improvements over the past year merit special mention. First, the mechanism that links the identifiers between Ensembl genes, transcripts and translations and their counterparts in external databases has been significantly improved and extended, including a new configuration system allowing us to appropriately address specific data types and relationships between external and Ensembl data. Second, we have expanded the automatic data quality checks that are vital to ensuring that the billions individual pieces of Ensembl data are as accurate as possible. There are now nearly 300 such tests that run in advance of each Ensembl release.
Comparative genomics
The protein tree calculation pipeline has evolved since last year with closer collaboration with the TreeFam project (http://www.treefam.org). TreeBeST software (http://treesoft.sourceforge.net) is used to both build a protein tree and reconcile it with the species tree. This reconciliation step allows us to call duplication and speciation events in the tree. Next, we check for dubious duplication events. These correspond to prediction where a duplication event is followed by a large number of gene loss events. Finally, we can infer paralogy and orthology relationships between the genes using the resulting protein tree.
Multiple genomic alignments are now calculated using Pecan (http://www.ebi.ac.uk/~bjp/pecan/) as it has been shown to be one of the best algorithms in terms of specificity and sensitivity (27). The new set of alignments includes the platypus genome. Each position in these alignments is further analysed to evaluate the level of evolutionary constraint using GERP as previously described (28). GERP also defines stretches of the Pecan alignments with a high level of conservation called constrained elements (Figure 1).
Data mining for comparative genomics
ComparaMart is a new data mining tool created to allow researchers to create intuitive queries against the Ensembl Compara multi-species database. ComparaMart uses the BioMart (6) data federation technology and provides a powerful, flexible tool to access a subset of the Compara data including predictions of homologues proteins and whole genome alignments.
As noted above, the Compara database stores results of genome-wide species comparisons calculated for each release. The ComparaMart database includes three main data sets: Ensembl homology, Ensembl pair-wise alignments and Ensembl multiple alignments. Through the ComparaMart interface, users may access the Ensembl homology data set to retrieve orthology or paralogy information for two species including various identifiers, homology descriptions, DNA/peptide sequences and peptide alignments. Additionally, the Ensembl homology data can also be linked to any Ensembl species-specific data sets to build more complex queries such as a list of all SNPs in human and mouse one-to-one orthologues. Specific data mining for pair-wise and multi-species whole-genome alignments are accessible through their respective data sets, although the multiple alignments data set includes only the constrained elements defined by GERP (28) from the Pecan alignments of 10 amniota vertebrates.
Outreach
Ensembl continuously tries to enhance the user experience and for this purpose we are in touch with our user community. This year we added video tutorials at http://www.ensembl.org/info/helpdesk/tutorials/index.html and continue to provide on-site courses on request. In an effort to gather information from Ensembl users and better understand how people use Ensembl, we recently conducted our second major user survey. More than 450 people responded primarily from Europe and North America. The results show overall satisfaction with Ensembl's tools and resources. For example, the most important aspects of Ensembl are accurate information (60% of respondents), followed by high-quality data visualization (41%), constant availability (36%), and good data mining tools (33%). Interestingly, the most common user concern was also related to data visualization, specifically the complexity of the Ensembl web interface. We are have already responded to several aspects of the survey and plan to make significant improvements to the web interface in 2008 to address the concerns raised.
FUTURE DIRECTIONS
The success of massively parallel sequencing technologies is a significant challenge for bioinformatics resources, although one that has been at least partially anticipated by Ensembl. We envision many ways this new technology will impact Ensembl over the coming year. We expect that resequencing data will be a significant part of Ensembl development over the next year and are working to scale our resequencing and variation resources appropriately. The sequencing technologies have likely made whole genome tiling array analysis obsolete (at least for ChIP) and we are adapting our functional genomics database for ChIP-seq analysis support. We anticipate continued enhancements of the Ensembl regulatory build as new genome-wide data sets become available through projects such as ENCODE. Finally we expect that new transcriptomics data sets will help us guide the Ensembl gene build both in terms of improving currently supported species and mapping transcription in newly sequenced genomes.
ACKNOWLEDGEMENTS
The Ensembl project receives primary funding from the Wellcome Trust. Additional funding is provided by EMBL, NHGRI, NIH-NIAID, BBSRC, MRC and the European Union. We acknowledge those researchers and organizations (especially Greg Crawford, Martin Hirst and the STAR Consortium) that have provided data to Ensembl prior to publication under the understandings of the Fort Lauderdale meeting discussing Community Resource Projects. We thank all of the users of our website and other resources, and those who have provided useful feedback though our mailing list. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasprzyk A, Keefe D, Smedley D, London D, Spooner W, Melsopp C, Hammond M, Rocca-Serra P, Cox T, et al. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potter SC, Clarke L, Curwen V, Keenan S, Mongin E, Searle SM, Stabenau A, Storey R, Clamp M. The Ensembl analysis pipeline. Genome Res. 2004;14:934–941. doi: 10.1101/gr.1859804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rayner TF, Rocca-Serra P, Spellman PT, Causton HC, Farne A, Holloway E, Irizarry RA, Liu J, Maier DS, et al. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. BMC Bioinformatics. 2006;7:489. doi: 10.1186/1471-2105-7-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MGC Project Team. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng P, Wei CL, Sung WK, Chiu KP, Lipovich L, Ang CC, Gupta S, Shahab A, Ridwan A, et al. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nat. Methods. 2005;2:105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- 15.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 16.Ruan Y, Ooi HS, Choo SW, Chiu KP, Zhao XD, Srinivasan KG, Yao F, Choo CY, Liu J, et al. Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using Paired-End diTags (PETs) Genome Res. 2007;17:828–838. doi: 10.1101/gr.6018607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefranc MP, Giudicelli V, Kaas Q, Duprat E, Jabado-Michaloud J, Scaviner D, Ginestoux C, Clément O, Chaume D, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2005;33:D593–D597. doi: 10.1093/nar/gki065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mardis ER. Anticipating the 1,000 dollar genome. Genome Biol. 2006;7:112. doi: 10.1186/gb-2006-7-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bentley DR. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunningham F, Rios D, Griffiths M, Smith J, Ning Z, Cox T, Flicek P, Marin-Garcin P, Herrero J, et al. TranscriptSNPView: a genome-wide catalog of mouse coding variation. Nat. Genet. 2006;38:853. doi: 10.1038/ng0806-853a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birney E, Andrews D, Caccamo M, Chen Y, Clarke L, Coates G, Cox T, Cunningham F, Curwen V, et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prlic A, Down TA, Kulesha E, Finn RD, Kahari A, Hubbard TJ. Integrating sequence and structural biology with DAS. BMC Bioinformatics. 2007;8:333. doi: 10.1186/1471-2105-8-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stabenau A, McVicker G, Melsopp C, Proctor G, Clamp M, Birney E. The Ensembl core software libraries. Genome Res. 2004;14:929–933. doi: 10.1101/gr.1857204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margulies EH, Cooper GM, Asimenos G, Thomas DJ, Dewey CN, Siepel A, Birney E, Keefe D, Schwartz AS, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper GM, Stone EA, Asimenos G, NISC Comparative Sequencing Program. Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Regha K, Sloane MA, Huang R, Pauler FM, Warczok KE, Melikant B, Radolf M, Martens JH, Schotta G, et al. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol. Cell. 2007;27:353–366. doi: 10.1016/j.molcel.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]