Abstract
Many Microbe Microarrays Database (M3D) is designed to facilitate the analysis and visualization of expression data in compendia compiled from multiple laboratories. M3D contains over a thousand Affymetrix microarrays for Escherichia coli, Saccharomyces cerevisiae and Shewanella oneidensis. The expression data is uniformly normalized to make the data generated by different laboratories and researchers more comparable. To facilitate computational analyses, M3D provides raw data (CEL file) and normalized data downloads of each compendium. In addition, web-based construction, visualization and download of custom datasets are provided to facilitate efficient interrogation of the compendium for more focused analyses. The experimental condition metadata in M3D is human curated with each chemical and growth attribute stored as a structured and computable set of experimental features with consistent naming conventions and units. All versions of the normalized compendia constructed for each species are maintained and accessible in perpetuity to facilitate the future interpretation and comparison of results published on M3D data. M3D is accessible at http://m3d.bu.edu/.
INTRODUCTION
Microarrays, once a selectively used expensive tool, have become increasingly common due to their falling costs and increased credibility over the past 10 years. In contrast to the bulk of DNA sequencing, which has been taken over by large centers that automatically submit sequencing reads to centralized databases (e.g. GenBank), the majority of microarray expression data is still generated by smaller laboratories addressing particular biological questions.
Given the diversity of expression possibilities in the cell and the stochastic nature of transcription and of microarrays themselves, previous studies have found computational analysis of large sets of microarrays (compendia) to be a powerful means of identifying strong biological signals between genes and across conditions (1–4). Historically these compendia have been generated as large internally controlled projects from a single laboratory, often excluding smaller datasets from independent laboratories. Yet, the many small microarray datasets generated worldwide represent a large and underutilized resource for genome-scale analyses such as compound mode of action identification (5) and network inference (6,7).
The creators of the GEO database at NCBI (8) and the ArrayExpress database at EBI (9) have sought to address this opportunity by providing a central repository of expression data for large and small laboratories. While valuable first initiatives, the GEO and ArrayExpress databases are not yet structured in a way that facilitates efficient exploration or analysis of the data. Four main obstacles exist: first, submitting microarray datasets to repositories is more difficult than submitting sequences to GenBank—the data itself is more complicated, requiring submission formats that are beyond the means of many non-computational researchers. Thus a significant number of published array datasets have not been deposited. This problem has been addressed to some degree as more journals require submission of microarray data to GEO or ArrayExpress.
The second obstacle is the presence of platform-specific biases in expression data due to the use of many different microarray platforms in a compendium. These biases obfuscate the interpretation of the integrated dataset. For dual-channel arrays, the situation is often further complicated by the lack of a single physiological reference condition used across all arrays in the platform. This lack of uniform reference prohibits some types of computational analyses. Hence, the first step in the analysis of an array compendium is often to segregate data into sets with a uniform reference condition and a consistent array type. This time-consuming step often reduces the compendium to a far less expansive dataset.
The third obstacle is the lack of uniformity in the format of expression data, even within a single expression platform. Various software algorithms are available for preprocessing and normalizing the raw microarray intensity values. The data deposited in GEO and ArrayExpress does not necessarily employ a uniform preprocessing approach, nor is the raw intensity data always provided with the deposits. Thus, end-user performed preprocessing and normalization is precluded.
The fourth obstacle is the incompleteness and inconsistency in the curation of metadata describing the details of each experimental condition. Each expression profile run for a given species can have a different genetic background, media, growth conditions and any number of chemicals, which might have an effect on the cell's expression. Such data is fundamental to the meaningful interpretation of expression data. Even when provided, this metadata is found as unstructured prose in the database deposit or in the methods sections of each publication. Ideally, this metadata would be collected in a computable format with uniform units across all laboratories. Although standards like MIAME (10) promote the human interpretation of experimental conditions, the standard is unevenly applied and it does not facilitate computational analysis.
To address the latter three of these problems, we have constructed the Many Microbe Microarrays Database (M3D). M3D currently contains over 1000 microarrays for Escherichia coli (507), Saccharomyces cerevisiae (530) and Shewanella oneidensis (14), all of which were collected and combined from individual investigators, GEO (8), ArrayExpress (9) and ASAP (11). To avoid problems with platform-specific biases, M3D contains only single-channel Affymetrix microarrays. The expression data is uniformly normalized to enable web-based or offline (via a database dump) analysis without further user-dependent normalization. This facilitates analysis of the data across all laboratories and conditions, even by non-expert users. A set of web-based browsing and analysis tools is provided to facilitate efficient interrogation of the dataset without extensive computational skills. Raw intensity data files are also provided for all datasets for expert users. Importantly, experimental metadata in M3D is human curated from each microarray publication—converting each chemical and growth attribute into a structured and computable set of experimental features with consistent naming conventions and units. Finally, all versions of the database builds are maintained and accessible in perpetuity on the website to facilitate the future interpretation and comparison of results published on M3D data.
The various attributes of M3D—comprehensive data and metadata, uniform normalization, access to raw data dumps, a computable structure, versioning of the database and web-based analysis tools—facilitate both efficient human interrogation of the dataset and machine-based computational analysis. Moreover, the consistency and uniformity of the dataset facilitates downstream comparison of results and findings based on the dataset.
SINGLE-PLATFORM, SINGLE-CHANNEL, UNIFORMLY NORMALIZED
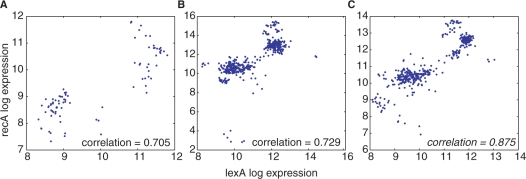
Large microarray depositories like GEO and ArrayExpress focus on the archiving of expression data as used in specific publications. These archives play an essential role in biological science by allowing transparent replication of microarray analyses by other researchers. Experimenters using the same array platform often use different normalization methods for their analyses, so that data downloaded from different projects on GEO or ArrayExpress are unlikely to be directly comparable. GEO at NCBI provides GEO DataSets to alleviate this problem. A GEO DataSet contains a collection of biologically and statistically comparable microarray samples processed using the same platform. Unfortunately, there is a significant delay between when a sample is submitted to GEO and when it is available as a GEO DataSet. Only one-fifth of the number of samples in M3D were available from GEO DataSets (Figure 1A and B).
Figure 1.
All of the available E. coli Affymetrix Antisense2 expression data for the transcription factor lexA and its known target recA were downloaded from NCBI GEO Profiles (A) and from M3D compendium E_coli_v3_Build_1 (B and C). NCBI GEO Profile data is derived from NCBI GEO DataSets that contain only a subset of the data in GEO, therefore many more samples were available for plotting from M3D (445) than from GEO (85). The correlation between lexA and its known target was higher when the raw data was uniformly normalized with RMA (C) rather than normalizing each microarray individually with MAS5 (A and B).
We have initially chosen to include only single-channel Affymetrix microarrays in M3D. The photolithography process used by Affymetrix allows all laboratories to start with a very consistent substrate for hybridization. In addition, the single-channel design eliminates the need for a common reference condition for all arrays. Thus, in contrast to two-color array designs, data from different laboratories and projects can be integrated without artifacts due to an inconsistent reference condition. The remaining systematic biases in the Affymetrix platform are due to researcher-specific differences in the RNA preparation and hybridization protocols. However, when the raw probe-level microarray data (CEL files) are normalized as a group with RMA (12), we find that these systematic researcher biases are small relative to the biological changes that occur across experimental conditions (7). In addition, the RMA normalized data tends to have higher correlation between the expression of transcription factors and their known targets (Figure 1B and C).
To employ the RMA normalization approach in M3D, all expression profiles for a particular array design (e.g. the E. coli Antisense 2 array) are collected, uniformly normalized and deposited as a ‘build’. Periodically, we add new expression profiles for a particular array design, renormalize all data, and release a new ‘build’. This ensures that all experiments in any build are uniformly normalized and comparable across conditions. The renormalization process may result in small changes in the expression values of all profiles. Thus, all builds are labeled with a version number that references the underlying mysql schema of the database and a build number that denotes the particular set of microarray data (e.g. E_coli_v3_Build_2 uses mysql schema version 3 and is the second compendium built for E. coli). Builds are maintained in perpetuity. This system, like the build system used by the human genome assembly, allows computational researchers to specify the exact dataset used for a particular analysis.
CURATED, COMPUTABLE EXPERIMENTAL METADATA
The experimental condition information underlying each microarray sample is the most under-utilized aspect of compendia collected from multiple disparate sources. In scientific language there are typically multiple units that can be used to describe a particular aspect of an experiment. For example, the amount of glucose added to a media can be described in weight/volume, as a percent solution, or using molarity. To promote large-scale analyses of the relationship between experimental conditions and the expression values of each gene, we provide the quantitative and qualitative features of each experimental condition cataloged in a consistent framework suitable for computation. We use human curation to convert the condition metadata in each publication into consistent units and naming conventions, and we use computer validation to provide data integrity.
BULK DOWNLOADS
To facilitate large-scale computational analyses of compendium data, we provide bulk downloads of the normalized expression data in M3D. For each build, we provide separate files containing normalized data for all genes, all genes + intergenic regions, and all genes + intergenic regions + control probes. We also provide flat files containing the gene names, probe set names and curated experimental condition information. In addition, we provide the raw CEL files as a tar archive for researchers interested in using or developing other normalization methods.
ONLINE ANALYSIS, VISUALIZATION AND CUSTOM DATA DOWNLOADS
For more targeted analysis and data exploration, M3D allows the flexible construction, visualization and download of custom datasets. Users can select any subset of the experiments in M3D using checkboxes or by selecting ‘projects’ that represent larger groups of experiments (typically a project is the set of microarrays available in a single publication). Similarly, users can choose a subset of genes by typing or uploading a list of gene and/or probe names. Genes can also be selected by differential expression as measured by t-test, z-test or fold change (e.g. choose all genes with a significant expression change between experiments A,C,E versus B,D,F,G as measured by a t-test with a user-chosen significance threshold).
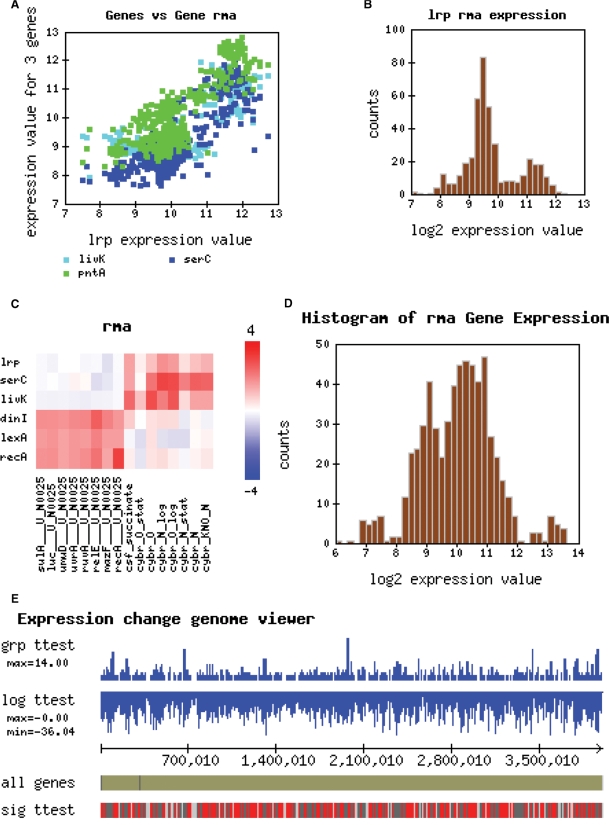
Once a user selects a set of genes and experiments, the data can be downloaded or visualized. Although, there are many existing general plotting tools and a few software visualization products dedicated to microarrays, it is often convenient to be able to choose a few conditions of interest, type in a few genes and see a quick plot of the data. M3D currently provides heat plots (with and without clustering), expression histograms (for individual genes and groups of genes), scatter plots and a genome browser for visualization of expression in a genome context (Figure 2) (13).
Figure 2.
Custom datasets constructed on M3D can be visualized with scatterplots (A), histograms of individual genes (B), heatplots (C), histograms of collections of genes (D) and in their genome context using a genome browser (E).
All browsing, analytical and download features of M3D are accessed from the same page, the Analysis page, on the website. This page guides the user step-by-step through the process of database selection, experiment selection, gene selection, visualization, analysis and download. At each step, a user's selections are saved in a cookie, enabling the user to return to and modify any prior selection without losing any other selections. The user can also select ‘Start Over’ from any point in the analysis to clear all selections. Context-specific help is provided on each page by mousing over the ‘[?]'symbol.
REMOTELY ACCESSIBLE VISUALIZATION
The power of the Internet resides in its interconnectivity. Biological databases like NCBI (14), Ecocyc (15) and RegulonDB (16) provide easy linking mechanisms so that other databases can automatically generate hyperlinks to their content. M3D provides a simple mechanism for generating links to M3D GenePages, which contain a basic set of plots for each gene. In addition all of the plots on M3D are generated using a simple URL syntax so that websites can easily include plots generated by M3D on their own sites. For example, strong correlation is often found between the expression of a transcription factor and its targets, a website cataloging known or predicted regulatory interactions might find it useful to provide a scatter plot of the transcription factor's expression versus its target gene's expression to allow users to see if expression data currently supports the regulatory interaction. The URL syntax for this and other plots can be found by clicking the help tab on the main menu of M3D.
UPDATE AND DATA SUBMISSION PROCEDURES
Raw CEL files are periodically collected from the ArrayExpress and GEO microarray databases. Upon accumulation of approximately 50 new chips for a particular species, all of the old and new microarrays are normalized together into a new compendium build. For researchers preferring to submit CEL files directly to M3D, we can generate a template submission to GEO, which the researcher can then edit as desired.
ACKNOWLEDGEMENTS
This research was supported by the Office of Science (BER), U.S. Department of Energy, Grant Nos. DE-FG02-04ER63803 and DE-FG02-07ER64388, the National Institute of General Medical Science, Grant No R01 GM078987, and the Joint NSF/NIGMS Mathematical Biology Program. Funding to pay the Open Access publication charges for this article was provided by the U.S. Department of Energy.
Conflict of interest statement. None declared.
REFERENCES
- 1.Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 2.Gardner TS, Faith JJ. Reverse-engineering transcription control networks. Phys. Life Rev. 2005;2:65–88. doi: 10.1016/j.plrev.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 4.Liao JC, Boscolo R, Yang YL, Tran LM, Sabatti C, Roychowdhury VP. Network component analysis: reconstruction of regulatory signals in biological systems. Proc. Natl Acad. Sci. USA. 2003;100:15522–15527. doi: 10.1073/pnas.2136632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.di Bernardo D, Thompson MJ, Gardner TS, Chobot SE, Eastwood EL, Wojtovich AP, Elliott SJ, Schaus SE, Collins JJ. Chemogenomic profiling on a genome-wide scale using reverse-engineered gene networks. Nat. Biotechnol. 2005;23:377–383. doi: 10.1038/nbt1075. [DOI] [PubMed] [Google Scholar]
- 6.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 7.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles – database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N, Lilja P, et al. ArrayExpress–a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 11.Glasner JD, Liss P, Plunkett G, III, Darling A, Prasad T, Rusch M, Byrnes A, Gilson M, Biehl B, et al. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 2003;31:147–151. doi: 10.1093/nar/gkg125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faith JJ, Olson AJ, Gardner TS, Sachidanandam R. Lightweight genome viewer: portable software for browsing genomics data in its chromosomal context. BMC Bioinformatics. 2007;8:344. doi: 10.1186/1471-2105-8-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salgado H, Gama-Castro S, Peralta-Gil M, Diaz-Peredo E, Sanchez-Solano F, Santos-Zavaleta A, Martinez-Flores I, Jimenez-Jacinto V, Bonavides-Martinez C, et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–D397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]