Abstract
Transcription factors (TFs) play key roles in controlling gene expression. Systematic identification and annotation of TFs, followed by construction of TF databases may serve as useful resources for studying the function and evolution of transcription factors. We developed a comprehensive plant transcription factor database PlantTFDB (http://planttfdb.cbi.pku.edu.cn), which contains 26 402 TFs predicted from 22 species, including five model organisms with available whole genome sequence and 17 plants with available EST sequences. To provide comprehensive information for those putative TFs, we made extensive annotation at both family and gene levels. A brief introduction and key references were presented for each family. Functional domain information and cross-references to various well-known public databases were available for each identified TF. In addition, we predicted putative orthologs of those TFs among the 22 species. PlantTFDB has a simple interface to allow users to search the database by IDs or free texts, to make sequence similarity search against TFs of all or individual species, and to download TF sequences for local analysis.
INTRODUCTION
Transcription factors (TFs) are important regulators to activate or repress the expression of coding or non-coding genes, through which they can further influence or control many biological processes. TRANSFAC collects ample information about animal transcription factors and their known binding cis-elements, with much less information about plant transcription factors (1). The completion of Arabidopsis thaliana genome sequencing made it the first model plant for transcription factor studies at the whole genome level (2). Several Arabidopsis TF online databases such as AtTFDB and RARTF are available over the Internet (3,4). In previous work, we have systematically predicted and annotated the transcription factors in Arabidopsis, rice and poplar based on their genome sequences, and constructed three distinct TF databases (5–7). Nevertheless, the requirement for an integrated and user-friendly plant transcription factor database is increasing, while the genomic sequencing of more and more plant species is underway. Riano-Pachon et al. (8) has constructed a database of transcription factors for five plant species and made the first attempt for construction of a comprehensive plant transcription factor database. The PlanTAPDB developed by Rensing and his colleagues is a comprehensive phylogeny-based resource of plant transcription associated proteins (9).
Here, we report a comprehensive plant TF database with 22 species (http://planttfdb.cbi.pku.edu.cn). In addition to the five species with complete genome sequences (Arabidopsis, rice, poplar, green alga and moss), we have also identified and annotated TFs based on the transcripts assembled by PlantGDB (10) for 17 plant species including crops, fruits, trees and other economically important plants. Detailed annotations were provided for each predicted TF. Furthermore, we have predicted TF orthologs in those species for comparative analysis and evolutionary studies. Both the sequences and annotation information for each identified TF are freely available for online access on the PlantTFDB website. We hope that PlantTFDB may become a useful resource for the research community, especially in the study of comparative genomics and transcription regulation.
DATA SOURCES AND METHODS
Data sources
Currently, PlantTFDB contains TFs identified in 22 species (Table 1). Genome sequences of Arabidopsis (A. thaliana), rice (Oryza sativa), poplar (Populus trichocarpa), green alga (Chlamydomonas reinhardtii) and moss (Physcomitrella patens) were downloaded from TAIR, TIGR and JGI. For the 17 species without available complete genome data, we downloaded the unique transcripts from the Plant Genome Database (PlantGDB, http://www.plantgdb.org/) (10). These plant unique transcripts (PUTs) were assembled by PlantGDB based on the mRNA and EST sequences. We applied the framefinder program in ESTate (Expressed Sequence Tag Analysis Tools Etc) package to predict the open reading frames and obtain protein sequences from these PUTs (http://www.ebi.ac.uk/~guy/estate/).
Table 1.
Basic information of 22 species and TFs in the current PlantTFDB
| Data source (Version)a | Name | Species | TFsb | TFs With Orthologsc | |
|---|---|---|---|---|---|
| TAIR (v6) | Arabidopsis | Arabidopsis thaliana | 2290 | 1346 | |
| JGI (v1.1) | Poplar | Populus trichocarpa | 2576 | 2042 | |
| TIGR (v4.0) | Rice | Oryza sativa (ssp. indica) | 2025 | 1763 | |
| Oryza sativa (ssp. japonica) | 2384 | 2124 | |||
| JGI (v1.1) | Moss | Physcomitrella patens | 1170 | 524 | |
| JGI (v3.0) | Green alga | Chlamydomonas reinhardtii | 205 | 64 | |
| PlantGDB (v155a) | Crops | Barley | Hordeum vulgare | 618 | 595 |
| Maize | Zea mays | 764 | 734 | ||
| Sorghum | Sorghum bicolor | 397 | 372 | ||
| Sugarcane | Saccharum officinarum | 1177 | 1157 | ||
| Wheat | Triticum aestivum | 1127 | 1074 | ||
| Fruits | Apple | Malus domestica | 1025 | 938 | |
| Grape | Vitis vinifera | 867 | 793 | ||
| Orange | Citrus sinensis | 599 | 541 | ||
| Trees | Pine | Pinus taeda | 950 | 644 | |
| Spruce | Picea glauca | 440 | 383 | ||
| Economic plants | Cotton | Gossypium hirsutum | 1567 | 1430 | |
| Potato | Solanum tuberosum | 1340 | 1243 | ||
| Soybean | Glycine max | 1891 | 1774 | ||
| Sunflower | Helianthus annuus | 513 | 435 | ||
| Tomato | Lycopersicon esculentum | 998 | 917 | ||
| Deervetch | Lotus japonicus | 457 | 434 | ||
| Medicago | Medicago truncatula | 1022 | 914 |
TAIR: The Arabidopsis Information Resource, http://www.arabidopsis.org/; TIGR: The Institute for Genomic Research, http://www.tigr.org/; JGI: DOE Joint Genome Institute, http://genome.jgi-psf.org/; PlantGDB: Plant Genome DataBase, http://www.plantgdb.org/.
The TF numbers of Arabidopsis and japonica rice are the number of gene models including alternative splicing.
The number of TFs of each species that has orthologs in all other species.
Plant TF HMM profiles
Transcription factors are always grouped as different families based on their DNA binding domains. Currently, 64 TF families have been characterized in plants (7). Among them, 48 families have hidden Markov Model (HMM) profiles in the Pfam database (v20.0) (11), while the remaining 16 families do not have available HMM profiles since they either were newly identified or only had a few members. To build the HMM profiles of these 16 families, we took their protein sequences from the previous TF databases of Arabidopsis, rice and poplar (5–7) and performed multiple sequence alignment. Then, we manually refined the alignment results and kept only the regions representing the conserved DNA binding domain. Finally, we used the hmmbuild program in the HMMER package (http://hmmer.janelia.org/, v2.3.2) to build the HMM profiles for these 16 TF families.
TF identification
We applied the hmmsearch program in HMMER to search against the protein sequences of each species to predict TFs. Based on our previous experience and manual inspection, we took E-value 0.01 as the cutoff, which was widely adopted for HMMER search.
Many TFs have more than one DNA binding domains (2). For example, the B3 domain (PF02362) was presented in either ABI3-VP1 family or RAV subfamily of the AP2 family. We assigned TFs into the ABI3-VP1 family if they only possessed the B3 domain, otherwise to the AP2 family if they had both B3 and AP2 domains (2). We developed a rules-driven program to handle such issues. Detailed rules to categorize the TFs can be found in the PlantTFDB help page (http://planttfdb.cbi.pku.edu.cn/help.php).
TF annotation
To provide comprehensive information for the identified TFs, we made extensive annotations on both the family and gene levels. For each TF family, a brief introduction and key reference were listed in the family page. BLAST search was performed against well-known public databases such as UniProt, RefSeq, EMBL and TRANSFAC. Putative functional domains were identified and annotated by InterProScan, and Gene Ontology annotations were further extracted. In addition, the expression profiles collected from UniGene EST/cDNA information were available for all putative TFs.
Putative ortholog annotation
To predict putative orthologous relationship of TFs among these species, we used the BLAST score ratio (BSR) method, which had been widely adopted by ENSEMBL (http://www.ensembl.org/) and other studies (12). An all-against-all BLASTP search with a strict cutoff E-value <1E−20 was performed, and the BSR value was calculated for each hit. After comparing results at different BSR value, we chose the BSR value ≥0.4 as the cutoff and we retrieved the top sequences in a species with the largest BSR value as the putative ortholog(s).
DATABASE CONSTRUCTION AND WEB INTERFACE
We used MySQL as the database management system and designed a uniform database structure for most of the species except for Arabidopsis, rice and poplar. Each species has its own separate database and annotations against EMBL, UniProt, Gene Ontology, RefSeq, TRANSFAC and UniGene were stored in individual tables.
PlantTFDB has a user-friendly entry point for each species. We kept the previously constructed database interface of Arabidopsis, rice and poplar and developed a uniform web interface for the 19 newly added species (Figure 1). A uniform text query interface for each species was designed. BLAST search against all or individual species was provided. All the sequences and ortholog information are available through the download page. Users can click the TF ID to activate the TF annotation information page with detailed annotations (Figure 1). In addition, putative orthologs among other species can also be found for each TF.
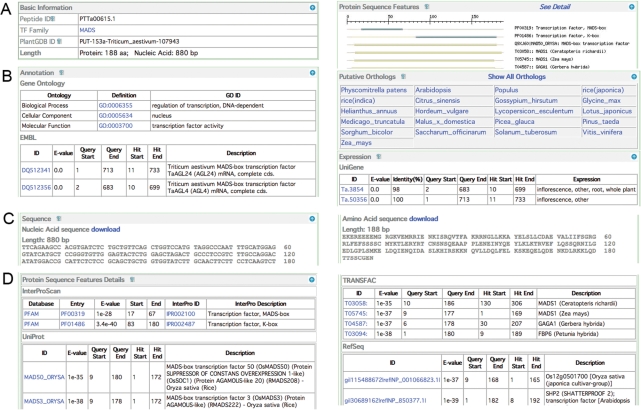
Figure 1.
The annotation information of a typical entry of the PlantTFDB showing the rich annotation of a bread wheat MADS box transcription factor (PTTa00615.1). The annotation contains four major categories: (A) Basic information; (B) Annotation; (C) Sequence; (D) Protein sequence feature details. Some of the annotations were tailored since the screen dump of the actual web page is too large to fit. The actual layout and content of the web page could be slightly different, since we keep developing and updating the database with new data available.
DISCUSSION
Protein sequences from PUTs
The PUT sequences assembled from transcripts may have insertion/deletion sites disrupting the open reading frames. We made TBLASTN against these PUTs using Arabidopsis proteins as query sequences and observed that more than half of them had frame shifts. Therefore, we used the framefinder program to obtain the protein sequences of the PUTs of 17 species.
Evaluation of self-built HMM profiles
Based on the multiple sequence alignment results of known members in Arabidopsis, rice and poplar, we built HMM profiles for DNA binding domains of 16 families, which did not have HMM profiles in the Pfam database (v20.0). We obtained the same hits from HMMER search against Arabidopsis proteins for CCAAT-HAP2 and MBF1 using the HMM profiles we built and the new HMM profiles added in Pfam (v22.0).
Evaluation of TF identification accuracy
To estimate the accuracy and reliability, we applied our pipeline to 10 well-annotated families in Arabidopsis. We measured the sensitivity and the specificity of our approach using the same approach described by Iida et al. (4) and Riano-Pachon et al. (8). Our results showed that the sensitivity and specificity of eight families were greater than 0.95, the sensitivity of two families were close to 0.90 and the specificity of one family was 0.935. This suggested that the approach we used had reasonable performance with acceptable accuracy.
Ortholog prediction
We used the widely-adopted BSR method to predict orthologs (12). To find an appropriate parameter, we made BLAST search against japonica rice to find orthologs of each Arabidopsis TF with six different BSR cutoffs (0.3, 0.33, 0.35, 0.4, 0.45 and 0.5) and compared the hit number, coverage and identity of the BLAST hits. Finally, we chose the BSR 0.4 as the cutoff of BLAST hits, which was relatively strict with an average coverage >80% and identity ∼60%. Based on these cutoffs, we chose the top sequences in a species with the largest BSR value as the putative ortholog(s).
CONCLUSION
PlantTFDB is our attempt for constructing a comprehensive plant transcription factor database with all currently available genome and transcript sequences. We will continue to add more species and new annotations when their sequence data become available. The extensive annotation of each specific TF family in 22 species and the information of orthologs among these species may facilitate the study of transcription regulation and the evolution of plant TFs.
ACKNOWLEDGEMENTS
This study was supported by grants from NSFC (90408015), 973 (2003CB715900), 863 (2006AA02Z334) and High-Tech Platform. Funding to pay the Open Access publication charges for this article was provided by the MOE grant to Kun He.
Conflict of interest statement. None declared.
REFERENCES
- 1.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 3.Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics. 2003;4:25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K. RARTF: database and tools for complete sets of Arabidopsis transcription factors. DNA Res. 2005;12:247–256. doi: 10.1093/dnares/dsi011. [DOI] [PubMed] [Google Scholar]
- 5.Zhu QH, Guo AY, Gao G, Zhong YF, Xu M, Huang M, Luo J. DPTF: a database of poplar transcription factors. Bioinformatics. 2007;23:1307–1308. doi: 10.1093/bioinformatics/btm113. [DOI] [PubMed] [Google Scholar]
- 6.Gao G, Zhong Y, Guo A, Zhu Q, Tang W, Zheng W, Gu X, Wei L, Luo J. DRTF: a database of rice transcription factors. Bioinformatics. 2006;22:1286–1287. doi: 10.1093/bioinformatics/btl107. [DOI] [PubMed] [Google Scholar]
- 7.Guo A, He K, Liu D, Bai S, Gu X, Wei L, Luo J. DATF: a database of Arabidopsis transcription factors. Bioinformatics. 2005;21:2568–2569. doi: 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- 8.Riaño-Pachón DM, Ruzicic S, Dreyer I, Mueller-Roeber B. PlnTFDB: an integrative plant transcription factor database. BMC Bioinformatics. 2007;8:42. doi: 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardt S, Lang D, Reski R, Frank W, Rensing SA. PlanTAPDB, a phylogeny-based resource of plant transcription-associated proteins. Plant Physiol. 2007;143:1452–1466. doi: 10.1104/pp.107.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Q, Lawrence CJ, Schlueter SD, Wilkerson MD, Kurtz S, Lushbough C, Brendel V. Comparative plant genomics resources at PlantGDB. Plant Physiol. 2005;139:610–618. doi: 10.1104/pp.104.059212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasko DA, Myers GS, Ravel J. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics. 2005;6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]