Abstract
Selenoproteins are a diverse group of proteins usually misidentified and misannotated in sequence databases. The presence of an in-frame UGA (stop) codon in the coding sequence of selenoprotein genes precludes their identification and correct annotation. The in-frame UGA codons are recoded to cotranslationally incorporate selenocysteine, a rare selenium-containing amino acid. The development of ad hoc experimental and, more recently, computational approaches have allowed the efficient identification and characterization of the selenoproteomes of a growing number of species. Today, dozens of selenoprotein families have been described and more are being discovered in recently sequenced species, but the correct genomic annotation is not available for the majority of these genes. SelenoDB is a long-term project that aims to provide, through the collaborative effort of experimental and computational researchers, automatic and manually curated annotations of selenoprotein genes, proteins and SECIS elements. Version 1.0 of the database includes an initial set of eukaryotic genomic annotations, with special emphasis on the human selenoproteome, for immediate inspection by selenium researchers or incorporation into more general databases. SelenoDB is freely available at http://www.selenodb.org.
INTRODUCTION
The identification of selenoprotein genes in sequenced genomes is particularly difficult and illustrates one of an increasing number of exceptions to our standard theory of the gene. In these proteins, the rare selenium-containing amino acid selenocysteine (Sec), chemically similar to cysteine (Cys), is cotranslationally incorporated in response to an in-frame UGA codon (1). Almost without exception, gene annotation pipelines rely on the standard termination codons UAA, UAG and UGA to identify open reading frames (ORFs) and predict coding exons. The dual role for the UGA codon within selenoprotein genes confounds both computational gene predictors and human curators, and results in the misannotation of selenoprotein gene structure in the majority of genome projects and databases. Furthermore, because the Sec residue is known to be located in the active site of most characterized selenoproteins, truncated predictions of these genes result in poorly characterized protein functions. To date, the majority of correctly annotated selenoproteins are mammalian mRNAs in GenBank entries submitted by experimental researchers, therefore lacking a genomic context (2). The selenoprotein annotations in the Recode database (3) are based on such mRNA entries. Multispecies genomic annotation systems like Ensembl annotate selenoproteins with different degrees of reliability between species and database releases (4), although the Ensembl-based Vertebrate Genome Annotation (VEGA) project annotates selenoprotein genes, except for the SECIS element, increasingly well (5). Overall, few selenoproteins are correctly and fully annotated in current genomic databases.
The interactions of several cis- and trans-acting factors are required in order to dynamically recode the UGA codon as Sec. The SElenoCysteine Insertion Sequence (SECIS) is the major cis-element (6,7). This sequence, found in the 3′ untranslated region (UTR) of selenoprotein genes in eukaryotes and archaea, and in the coding region in bacteria, forms a necessary stem-loop structure that recruits required trans-elements (8). SECIS structures differ among the three domains of life and are sometimes annotated in mRNA entries but rarely in genomic sequences and databases. Trans-acting proteins and other proteins related to selenium metabolism are usually not selenoproteins and, therefore, are predicted with average quality by gene prediction programs.
Recently, a number of computational approaches have been developed that have greatly contributed to the characterization of selenoproteins in many eukaryotic and prokaryotic genomes. Specific methods have been developed to identify selenoproteins by predicting SECIS elements (9–11). SECIS predictions, in turn, have been used to instruct modified gene prediction algorithms to ignore UGA codons as terminators when searching for new selenoprotein genes (12–14). In addition, comparative sequence analysis methods have proven very powerful in uncovering novel selenoproteins (15–17). The combination of these methods produces robust estimates of the size of a species selenoproteome (13,18). These computational approaches, together with standard homology methods of gene annotation and extensive manual curation, underpin the annotation process in SelenoDB.
However, selenium and selenoprotein biology remains a challenging field. First, the number, functional diversity and phylogenetic distribution of selenoproteins in nature are not precisely known. In fact, an ever growing number of selenoproteins is being discovered as sequencing of genomes progresses. Second, although selenoproteins with characterized functions are enzymes involved in redox reactions, the majority of selenoproteins have not been functionally characterized (19). The growing body of evidence linking selenium deficiency to a number of pathologies (20), makes current research on selenoprotein functions particularly relevant. Third, the atypical translational features of Sec make selenoproteins an outstanding research model of the translational process. Fourth, the phylogenetic and functional relationships among selenoprotein families are not fully understood, a necessary step to investigate the evolution of Sec usage in proteins. Fifth, the unclear, although possibly low (Castellano, unpublished data), extent of exchangeability between Sec and cysteine (Cys) (21–24), the uneven and dynamic use of these residues among species (14,17) and the uncertain origin of Sec in the genetic code make selenoprotein evolution an exciting problem. SelenoDB, version 1.0, is a long-term project specifically developed to assist the study of selenoproteins by providing high-quality annotations of selenoenzymes and Cys-containing homologs.
DATABASE CONSTRUCTION
Sequence identification
Three different types of sequences serve as the source for the annotation of selenoproteins and homologous sequences in SelenoDB: (i) Genomes (assemblies or traces); (ii) Transcripts (cDNAs or ESTs) and (iii) Proteins (full-length or fragments).
The identification of the best sequence available for annotation for a particular selenoprotein and species follows a comparative approach. Blast (25) and HMMER (26) programs are used to identify homologous sequences to human and other species selenoproteins. The search proceeds in this order: (i) genomic sequences from an annotated assembly from Ensembl or other major databases; (ii) full-length transcripts (cDNAs) with RefSeq (27), GenBank or TIGR (28) entries; (iii) partial transcripts (cDNAs or ESTs) with RefSeq, GenBank or TIGR entries; (iv) full-length proteins with UniProt (29) entries; (v) partial proteins with UniProt entries and (vi) from the literature.
Sequence feature annotation
Genes and other sequence features are annotated with one or more of the following complementary approaches:
- Comparative gene prediction followed by manual curation. This method is used to annotate through sequence homology known selenoproteins and related genes in new genomes. It is the preferred method of annotation in SelenoDB and can be divided into:
- Transcript homology with spidey (32) or exonerate. This approach is used to annotate non-coding regions in a gene.
Gene structures can be further refined with ab initio gene prediction methods using a modified version of geneid for selenoproteins (12,33) followed by manual curation. This method is used to annotate or reannotate known selenoproteins genes based on new sequence evidence (e.g. EST data).
SECIS prediction using SECISearch (13) followed by manual curation. This method is used to annotate SECIS elements in known selenoprotein sequences. This program is currently used to annotate eukaryotic selenoproteins. The future annotation of prokaryotic genes will rely, instead, on the bSECISearch program (34).
Note that third-party annotations are accepted in SelenoDB after standard quality controls. An easy step-by-step guide to format the data for SelenoDB is provided at the database site in the Documentation section. Researchers eager to have their gene of interest in the database are encouraged to contact us and/or follow the annotation instructions provided.
DATABASE ACCESS
The database can be accessed through its web server or through an anonymous MySQL account. The latter provides a user fluent in SQL the ability to interrogate the database in a customized manner (see Figure S1). In what follows, access to the database via the web interface is discussed.
Querying the database
Four search methods are available:
Simple keyword search: it is available, for convenience, at the top of each page in the site (Figure 1). It is a quick and easy way to search all or specific Feature Reports (Gene, Transcript, Protein, SECIS and others) with a single keyword. No keyword will search and match all Feature Reports in the database.
Simple species/family search: it is available, for convenience, at the bottom of each page in the site (Figure 1). It provides a fast link to Gene Reports for species selenoproteomes or selenoprotein families across species. The default search (All species and All families), will provide a list of all Gene Reports in the database.
Advanced search: t is available, for convenience, as a link at the top and bottom right of each page in the site under the name Advanced (Figure 1). It merges the two previous types of searches in a flexible and powerful searching schema that allows querying the Feature Reports by groups of common features. No selection will search all Feature Reports in the database.
Graphical search: it is available, for convenience, as a link at the top and bottom left of each page in the site under the name Chart (Figure 1). It is an integrated view of the annotated selenoproteomes by species and family in the database (Figure 1). Whole families or species together with single genes can be queried with a simple mouse click. The data is displayed in such a way to emphasize the dynamics of Sec/Cys usage throughout species and families. A click on the Chart will result in a list of all Gene Reports for the selected species, family or a particular gene family in one species.
Figure 1.
Graphical (Chart) search. Note the keyword and species/family search options at the top and bottom of the page, respectively. A click on a species, family or gene box leads to the corresponding query result page.
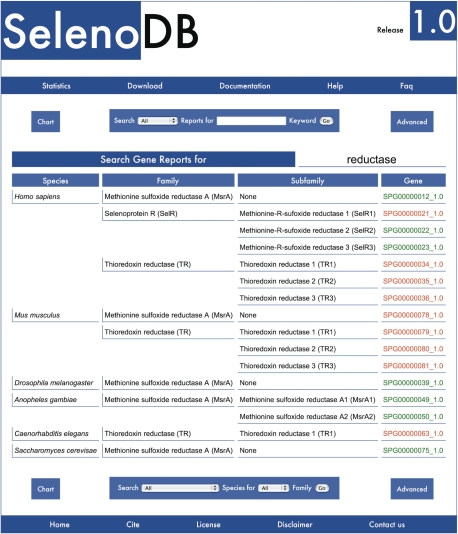
All search methods produce a list of matching Feature Reports links (Gene, Transcript, Protein and others) ordered by species, family and subfamily (Figure 2). Feature IDs are color coded: (i) selenoproteins in red; (ii) Cys-containing homologs in green; (iii) homologs bearing any other amino acid in the homologous Sec site in yellow and (iv) selenium machinery proteins or trans-acting proteins involved in the translational recodification of Sec residues in brown. The color code is maintained across all search results and Feature Reports, whenever a Feature ID is displayed.
Figure 2.
Query results from a keyword search on Gene Reports. Results are ordered by species, family and subfamily. Note the color coded Gene IDs links in the last column (red for selenoprotein and green for Cys-containing homolog).
Displaying features
The links in the Feature column of any search result page are used to display the corresponding Feature Report. In brief, the structure of the Feature Reports is the following:
Identity section. It provides information about the gene name and its accepted synonyms, family and subfamily names and their accepted synonyms, species and its taxonomical classification. In addition, internal and external (from other databases) Feature IDs are given. Internal Feature IDs are composed of: (a) a three-letter code, SPG, SPT, SPR, SPE, SPI, SPP, SPS and SEQ for gene, transcript, promoter, exon, intron, protein, SECIS and sequence, respectively; (b) an eight-digit number and (c) a version number to track changes in the annotation of sequence features. Version numbers will increase as the result of improved annotations due to: (1) higher quality sequences to replace the older annotated sequences; and (2) annotation errors reported by SelenoDB annotators or users. In any case, older annotated versions will remain available through the web interface.
Annotation section. In all Feature Reports, this section provides the annotation method (note the possibility to report annotation errors), orientation of the annotated Feature and displays the exonic structure of the annotated gene (Figure 3). In Gene Reports, a list of links to the corresponding Transcript, Promoter, Exon, Intron, Protein and SECIS Report is given. In all other Feature Reports, the Annotation section provides the sequence of the Feature of interest, e.g. the sequence of a particular exon in an Exon Report, the SECIS sequence in a SECIS Report or the transcript sequence in a Transcript Report (Figure 3). Both the gene structure plot and the displayed sequence in each Feature Report is color coded by feature type: (a) promoters in gray (note that promoter elements in the promoter regions are not yet provided due to the low quality of current computational prediction methods); (b) coding and non-coding exon regions (UTRs) in dark and light blue, respectively; (c) introns in black; (d) SECIS in brown and (e) Sec, Cys and other homologous residues in red, green and yellow, respectively. See Figure 3 for an example.
Sequence section. In Gene Reports, it provides information about the original source of the sequence used to annotate sequence features in SelenoDB and a link to the corresponding Sequence Report.
Figure 3.
Transcript Report of human SelO. Note the color coded gene structure and the spliced transcript sequence with sequences features colored accordingly. Other sequence views are one click away at links at the end of the sequence ID line.
Manipulating sequence features
The transcript sequence displayed in the Transcript Report can be manipulated in the following ways: (i) the full transcript can be spliced (Figure 3); (ii) the coding sequence (CDS) can be extracted from the transcript and (iii) the 5′ or 3′ untranslated regions (UTRs) can be extracted from the transcript. Annotated features in all sequences and subsequences are color coded (see Annotation section above for the color legend and Figure 3 for an example).
DATABASE STATISTICS
SelenoDB currently contains 81 genes from 8 different eukaryotic species, including the complete human, fly, mosquito and worm selenoproteomes. Selenoproteins from 20 different families are annotated. A detailed database statistics is available at the SelenoDB site.
DATABASE TECHNICAL DETAILS
SelenoDB is a relational database implemented in MySQL 5.0 with InnoDB as storage engine (see Figure S1 for a diagram of the database schema). The schema is designed to store non-standard genes with recoded codons, alternative translation initiation and termination sites, RNA secondary structures and other unusual features. Synchronization across future mirror sites will use the database server replication capacity. The selenodb_admin script in BASH manages the creation, removal, update and backup of the database. The SelenoDB web server runs on the open source software Apache 2.2.3 (embedded with mod_perl for optimal speed) with Perl 5.8.6 driving the database interface. The interface relies heavily on open web standards as cascading style sheets (CSS) 2.0, and the use of browsers complaint with current open standards is highly recommended. All the SelenoDB-specific software can be downloaded from the database site.
DATABASE LICENSE
SelenoDB is licensed under the GNU General Public License v2 or any later version.
DATABASE DOWNLOAD
In addition to the access to the data via a web server, the complete database package is available for download at the SelenoDB site. It includes the SQL database schema (see Figure S1), the script to create, remove, update and backup the database, a dump of the current database data and the web interface scripts. These software elements can also be downloaded separately.
FUTURE DIRECTIONS
SelenoDB is an ongoing collaborative project from experimental and computational labs. The long-term objectives of this project are twofold. First, to set up a reliable annotation framework (database and web interface) for selenoproteins, of which an initial but fully functional version is presented here. We would like to provide, in the future, sequence analysis tools (e.g. blast and hmmer) and the ability to include, in a wiki-like fashion, functional annotation of genes by selenium researchers. Second, to provide high-quality annotations of individual selenoproteins and complete selenoproteomes, of which an initial eukaryotic collection is presented in this work. The time-consuming nature of gene annotation and reannotation (error correction) and the focus on quality, makes us anticipate a slow but steady growth of the database. To help set priorities, researchers are encouraged to contact us for annotation instructions or to request specific genes to be annotated. In this direction, future plans include, besides the annotation of additional eukaryotic genomes, the annotation of prokaryotic selenoproteomes and of features of interest to selenium researchers, namely, alternative splice forms, protein domains and promoter elements (transcription factor binding sites) in promoter regions. Such comprehensive annotation could be the source of selenoprotein annotations for more general genomic databases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
S.C. thanks A.G. Clark and S.R. Eddy for time and resources to complete this project. This work was supported by National Institute of Health (NIH) grants DK47320 and DK52963 to M.J.B., grant BIO2006-03380 by the Spanish Ministry of Education and Biosapiens grant LSHG-CT-2003-503265 by the European Commission (FP6 Programme) to R.G. and NIH grant GM061603 to V.N.G. Funding to pay the Open Access publication charges for the article was provided by NIH DK47320 and DK52963.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, David L, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baranov PV, Gurvich OL, Hammer AW, Gesteland RF, Atkins JF. RECODE 2003. Nucleic Acids Res. 2003;31:87–89. doi: 10.1093/nar/gkg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashurst JL, Chen CK, Gilbert JG, Jekosch K, Keenan S, Meidl. P, Searle SM, Stalker J, Storey R, et al. The Vertebrate Genome Annotation (Vega) database. Nucleic Acids Res. 2005;33:D459–465. doi: 10.1093/nar/gki135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 7.Krol A. Evolutionarily different RNA motifs and RNA-protein complexes to achieve selenoprotein synthesis. Biochimie. 2002;84:765–774. doi: 10.1016/s0300-9084(02)01405-0. [DOI] [PubMed] [Google Scholar]
- 8.Allmang C, Krol A. Selenoprotein synthesis: UGA does not end the story. Biochimie. 2006;88:1561–1571. doi: 10.1016/j.biochi.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Kryukov GV, Kryukov VM, Gladyshev VN. New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J. Biol. Chem. 1999;274:33888–33897. doi: 10.1074/jbc.274.48.33888. [DOI] [PubMed] [Google Scholar]
- 10.Lescure A, Gautheret D, Carbon P, Krol A. Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J. Biol. Chem. 1999;274:38147–38154. doi: 10.1074/jbc.274.53.38147. [DOI] [PubMed] [Google Scholar]
- 11.Martin-Romero FJ, Kryukov GV, Lobanov AV, Carlson BA, Lee BJ, Gladyshev VN, Hatfield DL. Selenium metabolism in Drosophila: selenoproteins, selenoprotein mRNA expression, fertility, and mortality. J. Biol. Chem. 2001;276:29798–29804. doi: 10.1074/jbc.M100422200. [DOI] [PubMed] [Google Scholar]
- 12.Castellano S, Morozova N, Morey M, Berry MJ, Serras F, Corominas M, Guigó R. In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO Rep. 2001;2:697–702. doi: 10.1093/embo-reports/kve151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigó R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 14.Castellano S, Lobanov AV, Chapple C, Novoselov SV, Albrecht M, Hua D, Lescure A, Lengauer T, Krol A, et al. Diversity and functional plasticity of eukaryotic selenoproteins: identification and characterization of the SelJ family. Proc. Natl Acad. Sci. USA. 2005;102:16188–16193. doi: 10.1073/pnas.0505146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castellano S, Novoselov SV, Kryukov GV, Lescure A, Blanco E, Krol A, Gladyshev VN, Guigó R. Reconsidering the evolution of eukaryotic selenoproteins: a novel nonmammalian family with scattered phylogenetic distribution. EMBO Rep. 2004;5:71–77. doi: 10.1038/sj.embor.7400036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kryukov GV, Gladyshev VN. The prokaryotic selenoproteome. EMBO Rep. 2004;5:538–543. doi: 10.1038/sj.embor.7400126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Fomenko DE, Gladyshev VN. The microbial selenoproteome of the Sargasso Sea. Genome Biol. 2005;6:R37. doi: 10.1186/gb-2005-6-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taskov K, Chapple C, Kryukov GV, Castellano S, Lobanov AV, Korotkov KV, Guigó R, Gladyshev VN. Nematode selenoproteome: the use of selenocysteine insertion system to decode one codon in an animal genome? Nucleic. Acids Res. 2005;33:2227–2238. doi: 10.1093/nar/gki507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gromer S, Eubel JK, Lee BL, Jacob J. Human selenoproteins at a glance. Cell. Mol. Life Sci. 2005;62:2414–2437. doi: 10.1007/s00018-005-5143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium: Its Molecular Biology and Role in Human Health. 2nd edn. New York: Springer; 2006. [Google Scholar]
- 21.Axley MJ, Böck A, Stadman TC. Catalytic properties of an Escherichia coli formate dehydrogenase mutant in which sulfur replaces selenium. Proc. Natl Acad. Sci. USA. 1991;88:8450–8454. doi: 10.1073/pnas.88.19.8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berry MJ, Mai AL, Kieffer J, Harney JW, Larsen P. Substitution of cysteine for selenocysteine in type I iodothyronine deiodinase reduces the catalytic efficiency of the protein but enhances its translation. Endocrinology. 1992;131:1448–1852. doi: 10.1210/endo.131.4.1396330. [DOI] [PubMed] [Google Scholar]
- 23.Gromer S, Johansson L, Bauer H, Arscott LD, Rauch S, Ballou DP, Williams C.H., Jr, Schirmer RH, Arner ES. Active sites of thioredoxin reductases: why selenoproteins? Proc. Natl Acad. Sci. USA. 2003;100:12618–12623. doi: 10.1073/pnas.2134510100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HY, Gladyshev VN. Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases. PLoS Biol. 2005;3:e375. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 27.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y, Tsai J, Sunkara S, Karamycheva S, Pertea G, Sultana R, Antonescu V, Chan A, Cheung F, et al. The TIGR Gene Indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res. 2005;33:D71–D74. doi: 10.1093/nar/gki064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wheelan SJ, Church DM, Ostell JM. Spidey: a tool for mRNA-to-genomic alignments. Genome Res. 2001;11:1952–1957. doi: 10.1101/gr.195301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parra G, Blanco E, Guigó R. GeneID in Drosophila. Genome Res. 2000;10:511–515. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Gladyshev VN. An algorithm for identification of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics. 2005;21:2580–2589. doi: 10.1093/bioinformatics/bti400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.