Abstract
JASPAR is a popular open-access database for matrix models describing DNA-binding preferences for transcription factors and other DNA patterns. With its third major release, JASPAR has been expanded and equipped with additional functions aimed at both casual and power users. The heart of the JASPAR database—the JASPAR CORE sub-database—has increased by 12% in size, and three new specialized sub-databases have been added. New functions include clustering of matrix models by similarity, generation of random matrices by sampling from selected sets of existing models and a language-independent Web Service applications programming interface for matrix retrieval. JASPAR is available at http://jaspar.genereg.net.
INTRODUCTION
Computational analysis of regulatory properties of DNA is most often based on the use of matrix models describing binding preferences of transcription factors, or other DNA patterns. Such matrices are based on sets of known or inferred sites for a DNA-binding protein, and can be scanned over genomic sequences to predict novel binding sites (1,2). JASPAR is the most comprehensive open-access database holding such models. The heart of JASPAR is the JASPAR CORE sub database, holding curated, non-redundant matrix models from multi-cellular eukaryotes. The methodology for JASPAR CORE curation has been described previously (3). JASPAR CORE is now a standard resource in gene regulation bioinformatics and is used as a matrix set in a wide variety of other services [for instance (4–9)], and large-scale projects (10,11). Besides JASPAR CORE, the database contains several sub-databases (JASPAR Collections) holding matrix models produced by different methods and for different purposes (Table 1).
Table 1.
JASPAR databases
| Database | Number of models | Scope | Species coverage | When to use |
|---|---|---|---|---|
| JASPAR CORE | 138 | Curated, non-redundant matrix models | Multi-cellular eukaryotes | ‘Standard’ promoter analysis |
| JASPAR FAM | 11 | Familial ‘consensus’ patterns for major structural families of transcription factors | Multi-cellular eukaryotes | Matrix-to-matrix comparison and classification, or as prior knowledge for pattern finders |
| JASPAR PHYLOFACTS | 174 | Evolutionary conserved patterns in 5′ promoter regions | Multi-cellular eukaryotes | As a complement to JASPAR CORE for large-scale studies |
| JASPAR POLII | 13 | Core promoter element models | Multi-cellular eukaryotes | Core promoter analysis |
| JASPAR CNE | 233 | Motifs overrepresented in vertebrate highly conserved non-coding elements | Human | Analysis of regulatory content of long-range enhancers |
| JASPAR SPLICE | 6a | Splice sites | Humana | Splice site analysis |
aExpansion under way.
Here we present the recent JASPAR expansion, which includes a significant increase of the JASPAR CORE content and an addition of three new sub-databases focusing on core promoter patterns, splice sites and motifs detected in vertebrate highly conserved non-coding elements, respectively. In addition, we present several unique functional features in the web interface aimed at both casual and power users, including statistics on expected number of predictions each matrix will yield at several different thresholds in random sequences generated by three commonly encountered sequence background models, dynamic clustering of matrices by similarity and generation of random matrices using a selected set of matrices as background model.
RESULTS
Here we briefly describe the new data and functional features; more detailed descriptions are available at the documentation at the web site.
Expansion of JASPAR CORE
The JASPAR CORE database holds a curated set of transcription factor-binding profiles from multi-cellular eukaryotes: this is a unique feature with respect to databases of similar scope. We have extended JASPAR CORE with 15 new, high-quality profiles from recent experimental literature, increasing the total number of JASPAR CORE models to 138 (Table 1). In addition, annotation for all models in the database has been updated [e.g. to standard gene symbols from Entrez Gene (12)] and expanded. Prompted by user feedback, several existing matrices have been updated or corrected.
New sub-databases
Existing and new sub-databases within JASPAR and their specific features are described in Table 1. Since the last update, we have added three new sub-databases, which are briefly described below (see the web documentation for details):
JASPAR POLII
The large body of novel data pertaining transcription start sites (13,14) has triggered a new interest in computational studies of core promoters. The JASPAR POLII sub-database holds 13 known DNA patterns linked to RNA polymerase II core promoters, such as the Inr and BRE elements, each based on experimental evidence: each model must be constructed using five or more experimentally verified sites. An important difference to the transcription factor profiles in JASPAR CORE is that patterns here do not necessarily have a specified protein that binds them [See Ref. (15) for a review on core promoter patterns]. When possible, profiles were extended by 2 nt more than the core motif. We consistently report positions relative to the TSS as the position of 5′ and 3′ edge of the matrix.
JASPAR CNE
Highly conserved non-coding elements (CNEs) are a distinctive feature of metazoan genomes. Many of them can be shown to act as long-range enhancers that drive expression of genes that are themselves regulators of core aspects of metazoan development and differentiation. Since they act as regulatory inputs, attempts at deciphering the regulatory content of these elements have started (16–18). JASPAR CNE is a collection of 233 matrix profiles derived by Xie et al. (19) by clustering of overrepresented motifs from human conserved non-coding elements. While the biochemical and biological role of most of these patterns is still unknown, Xie et al. have shown that the most abundant ones correspond to known DNA-binding proteins, among them is the insulator-binding protein CTCF. These matrix profiles will be useful for further characterization of regulatory inputs in long-range developmental gene regulation in vertebrates.
JASPAR SPLICE
This small collection contains matrix profiles of human canonical and non-canonical splice sites, as matching donor:acceptor pairs. It currently contains only six highly reliable profiles (two canonical and four non-canonical) obtained from human genome (20). In the future, we shall include additional eukaryotic species, as well as new models for exonic splicing enhancers (ESE) and inhibitors (ESI).
Extended functionality
In addition to data extension, we have implemented a number of functional improvements in the web interface of the JASPAR database. These range from static statistics, such as expected number of hits on typical DNA sequence for any factor, to dynamic tools for similarity-based profile clustering and for generating random profiles based on a subset of known profiles.
Web service interface
The JASPAR database can now be reached remotely through a new Web Service interface. Current functionality includes retrieval of profiles by name, by identifier and by searching profile annotations. The purpose of providing an external application programming interface (API) is to simplify the utilization of JASPAR in distributed applications and in scientific workflows created in workflow editors like Triana (21), BPEL (http://www.bpelsource.com/) or Taverna (22). Other benefits include platform- and language-independent access, as well as constant up-to-date access to the database over time. The API is implemented as a WS-I compliant Web Service, identical to the technology used for the services made available through the EMBRACE Network of Excellence (www.embracegrid.info), and the Web Service technology chosen by the European Bioinformatics Institute (EBI) (23). Its basic usage is described in tutorials at the JASPAR web site. The WSDL describing this service can be found at: http://api.bioinfo.no/wsdl/JasparDB.wsdl. Further information about the Web Service, including example clients in Java and Python, is available on the Jaspar web site and in the WSDL file.
Expected predictions/base-pair statistics for all models
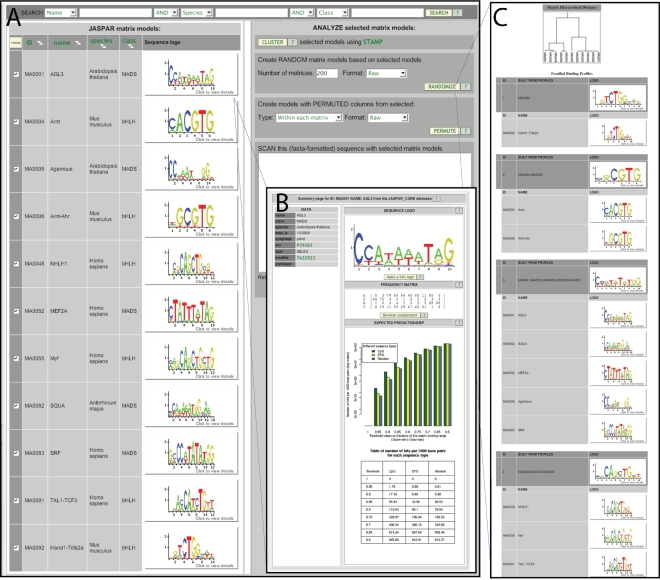
An important problem with genome-wide scanning with matrix models is the limited information content in a typical matrix, resulting in numerous spurious hits just due to sequence background (1,2). The number of false positives varies considerably between factors and also depends on what type of sequences that models are applied to, user-defined cutoffs and to a more limited extent on the type of scoring scheme used. For a first-glance assessment of the rate of spurious predictions of a given model, we apply the model to three distinct sequence sets: known promoters from the EPD database (24), CpG islands and randomly selected genomic DNA, respectively. For different score thresholds, we plot the mean number of hits per 1000 nt for each sequence set. The resulting bar plots are available for each JASPAR matrix (Figure 1).
Figure 1.
New features in the JASPAR database web interface. (A) A listing of matrices in the JASPAR-CORE database resulting from selection of MADS and bHLH-type factors. These models are used in the clustering analysis in panel C. (B) A pop-up window showing detail information on the MA0001 model, with expected predictions/bp statistics. (C) Dynamic clustering of selected profiles. At the top, a dendrogram describing the similarities of the input profiles is shown. Clusters of similar modes are merged into familial binding profiles, shown below. In this case, two larger clusters are produced, corresponding to bHLH and MADS type matrices. Two smaller clusters correspond to outliers in both groups.
Dynamic clustering by similarity and creation of familial binding profiles from a given profile subset
Many transcription factors bind similar targets and it is often helpful to cluster similar binding profiles to generate familial binding profiles—models describing a set of matrices (25). Part of this problem is matrix profile comparison and alignments, explored by several researchers (25–30). Recently, Mahony et al. (27,28) made a comprehensive study on alignments of matrices and construction of familial binding profiles, resulting in the STAMP tool, which is now used within JASPAR to cluster matrix models. Hierarchical clustering is performed on a selected set of matrices using the UPGMA algorithm with a Pearson Correlation Coefficient distance metric. Then the optimal number of clusters is selected using a log variant of the Calinski and Harabasz statistic [See Ref. (27) for details]. Finally, the clusters are partitioned and a familial binding profile is created for each cluster using iterative refinement (a multiple alignment method). An example is shown in Figure 1.
Dynamic random profile generation
In many computational studies, it is helpful to have a set of ‘random’ matrices. This is particularly true for assessment of distances between putative sites and reference points as transcription start sites, and also for matrix-to-matrix comparisons. In these cases, it is desired that the randomized matrices should share properties with the true matrix set—for instance having the same nucleotide content and/or the same general information content.
Within any JASPAR sub-database, users can select a subset of matrices, which will then be used to generate random matrices using one of two methods: (i) Permutations: Columns of the selected matrices are shuffled: either constrained to shuffling of columns within each matrix or between all selected matrices. (ii) Probabilistic sampling: This enables the users to generate random Position Frequency Matrices from selected profiles. In our model, each random column is sampled from a posterior distribution—a 4D Dirichlet mixture distribution. The posterior distribution has two contributions: a multinomial with counts of columns selected as in (i), and a Dirichlet mixture prior trained from all observed nucleotides in the JASPAR database. We assume that column positions are independent.
DISCUSSION
We have presented a significant update to the JASPAR database, including an expansion of the core database, three new sub-databases and many new utilities. The new web service interface enables easy interaction with scientific workflows and an increasing number of programming languages that support this technology. We project that the new features, together with the open-access policy, will further consolidate the JASPAR database as a standard resource in the field of gene regulation bioinformatics.
Towards a comprehensive set of models for most known transcription factors
The lack of models for the binding specificity of most transcription factors is a significant bottleneck for comprehensive computational analysis of genomes. Only a fraction of transcription factors have been characterized in enough detail to allow the construction of adequate models of their binding specificity. This problem is being solved in two principally different ways. First, tiling array approaches for measuring binding preferences en masse are being developed (31); these technologies show great promise and are expected to make their mark on the field in the near future. Second, a wealth of cis-regulatory elements, characterized in painstaking detail, is hidden in experimental literature; many of these sites are not included in any database. There is a growing awareness of this problem in the field, resulting in online open-access databases such as ORegAnno (32) and PAZAR (33), where one of the goals is to house expert-curated binding sites. We are currently developing services to enable cross-talk with these databases to enable matrix models built on curated sites that exceed a certain quality threshold. JASPAR, ORegAnno and PAZAR face the same challenge: to build models or sites, it is necessary to mine the literature, which inevitably means that the curators will miss many important studies. The only long-term solution would be a requirement by scientific journals for researchers to deposit protein–DNA interactions in public databases prior to publication, much in the same way as mRNAs must be submitted to Genbank (34). Part of such a system will be to establish a minimal standard for reporting these interactions, much like the MIAME standard (35) for microarray data. As before, JASPAR team is always prepared to incorporate new matrices and matrix sets provided by external contributors.
Data availability
All the data in JASPAR are available without any restrictions, either from the web interface, as flat files or through the Web service interface.
ACKNOWLEDGEMENTS
Thanks to Katsuya Shigesada for pointing out errors in matrix MA0002, Shaun Mahony and Panayiotis V. Benos for generously sharing the STAMP code and general helpfulness and Vladimir B. Bajic for kindly providing the frequency matrices for JASPAR SPLICE. E.V., M.-H.E.T., T.M., O.W., A.K. and A.S. were supported by a grant from the Novo Nordisk foundation to the Bioinformatics Center. I.P. was supported by a grant from Carlsberg Foundation (21-00-0680). J.C.B. was supported by EMBRACE—an EU Sixth Framework Network of Excellence. B.L. was supported by the Functional Genomics Programme (FUGE) of the Research Council of Norway, and a core grant from the Sars Centre. Funding to pay the Open Access publication charges for this article was provided by a grant from the Novo Nordisk Foundation and the Functional Genomics Programme of the Research Council of Norway.
Conflict of interest statement. None declared.
REFERENCES
- 1.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 2.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 3.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenhard B, Sandelin A, Mendoza L, Engstrom P, Jareborg N, Wasserman WW. Identification of conserved regulatory elements by comparative genome analysis. J. Biol. 2003;2:13. doi: 10.1186/1475-4924-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montgomery SB, Astakhova T, Bilenky M, Birney E, Fu T, Hassel M, Melsopp C, Rak M, Robertson AG, et al. Sockeye: a 3D environment for comparative genomics. Genome Res. 2004;14:956–962. doi: 10.1101/gr.1890304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marinescu VD, Kohane IS, Riva A. MAPPER: a search engine for the computational identification of putative transcription factor binding sites in multiple genomes. BMC Bioinformatics. 2005;6:79. doi: 10.1186/1471-2105-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aerts S, Van Loo P, Thijs G, Mayer H, de Martin R, Moreau Y, De Moor B. TOUCAN 2: the all-inclusive open source workbench for regulatory sequence analysis. Nucleic Acids Res. 2005;33:W393–W396. doi: 10.1093/nar/gki354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The ENCODE Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 12.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller F, Demeny MA, Tora L. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J. Biol. Chem. 2007;282:14685–14689. doi: 10.1074/jbc.R700012200. [DOI] [PubMed] [Google Scholar]
- 14.Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 15.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 16.Bailey PJ, Klos JM, Andersson E, Karlen M, Kallstrom M, Ponjavic J, Muhr J, Lenhard B, Sandelin A, et al. A global genomic transcriptional code associated with CNS-expressed genes. Exp. Cell Res. 2006;312:3108–3119. doi: 10.1016/j.yexcr.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 17.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 18.Pennacchio LA, Loots GG, Nobrega MA, Ovcharenko I. Predicting tissue-specific enhancers in the human genome. Genome Res. 2007;17:201–211. doi: 10.1101/gr.5972507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl Acad. Sci. USA. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chong A, Zhang G, Bajic VB. Information for the coordinates of exons (ICE): a human splice sites database. Genomics. 2004;84:762–766. doi: 10.1016/j.ygeno.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 21.Majithia S, Shields M, Taylor I, Wang I. Triana: a graphical web service composition and execution toolkit. Proceedings of the IEEE International Conference on Web Services; 2004. pp. 514–524. http://www.trianacode.org. [Google Scholar]
- 22.Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–W732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labarga A, Valentin F, Anderson M, Lopez R. Web services at the European bioinformatics institute. Nucleic Acids Res. 2007;35:W6–W11. doi: 10.1093/nar/gkm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmid CD, Perier R, Praz V, Bucher P. EPD in its twentieth year: towards complete promoter coverage of selected model organisms. Nucleic Acids Res. 2006;34:D82–D85. doi: 10.1093/nar/gkj146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandelin A, Wasserman WW. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J. Mol. Biol. 2004;338:207–215. doi: 10.1016/j.jmb.2004.02.048. [DOI] [PubMed] [Google Scholar]
- 26.Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- 27.Mahony S, Auron PE, Benos PV. DNA familial binding profiles made easy: comparison of various motif alignment and clustering strategies. PLoS Comput. Biol. 2007;3:e61. doi: 10.1371/journal.pcbi.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007;35:W253–W258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pietrokovski S. Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res. 1996;24:3836–3845. doi: 10.1093/nar/24.19.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schones DE, Sumazin P, Zhang MQ. Similarity of position frequency matrices for transcription factor binding sites. Bioinformatics. 2005;21:307–313. doi: 10.1093/bioinformatics/bth480. [DOI] [PubMed] [Google Scholar]
- 31.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep P.W., III., Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montgomery SB, Griffith OL, Sleumer MC, Bergman CM, Bilenky M, Pleasance ED, Prychyna Y, Zhang X, Jones SJ. ORegAnno: an open access database and curation system for literature-derived promoters, transcription factor binding sites and regulatory variation. Bioinformatics. 2006;22:637–640. doi: 10.1093/bioinformatics/btk027. [DOI] [PubMed] [Google Scholar]
- 33.Portales-Casamar E, Kirov S, Lim J, Lithwick S, Swanson MI, Ticoll A, Snoddy J, Wasserman WW. PAZAR: a framework for collection and dissemination of cis-regulatory sequence annotation. Genome Biol. 2007;8:R10. doi: 10.1186/gb-2007-8-10-r207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data in JASPAR are available without any restrictions, either from the web interface, as flat files or through the Web service interface.