Abstract
DIMA—the domain interaction map has evolved from a simple web server for domain phylogenetic profiling into an integrative prediction resource combining both experimental data on domain–domain interactions and predictions from two different algorithms. With this update, DIMA obtains greatly improved coverage at the level of genomes and domains as well as with respect to available prediction approaches. The domain phylogenetic profiling method now uses SIMAP as its backend for exhaustive domain hit coverage: 7038 Pfam domains were profiled over 460 completely sequenced genomes.Domain pair exclusion predictions were produced from 83 969 distinct protein–protein interactions obtained from IntAct resulting in 21 513 domain pairs with significant domain pair exclusion algorithm scores. Additional predictions applying the same algorithm to predicted protein interactions from STRING yielded 2378 high-confidence pairs. Experimental data comes from iPfam (3074) and 3did (3034 pairs), two databases identifying domain contacts in solved protein structures. Taken together, these two resources yielded 3653 distinct interacting domain pairs. DIMA is available at http://mips.gsf.de/genre/proj/dima.
INTRODUCTION
Conserved domains represent the building blocks of protein architecture. Many of the domains known today are re-used in a variety of different proteins in a modular fashion, thus conferring a large range of structural and functional features to their host proteins. The problem of biological annotation, formal description and classification of these domains has been addressed by several groups leading to important resources such as SMART (1), BLOCKS (2), PFAM (3) and the integration endeavor InterPro (4).
While the majority of conserved domains are mainly characterized by their biochemical activity or structural importance, a significant fraction represents adapters for physical binding. Examples include the well-described SH3, WW and PDZ domains (5), which are present in large numbers of functionally unrelated proteins, serving as universal interaction modules. Numerous approaches to the problem of systematically describing known domain interactions and identifying yet unknown interaction domains have been proposed (6–10). The PFAM database has added a domain interaction resource called iPFAM to their site that describes protein domains found to engage in physical contact (11). Known PFAM domains were matched to intra- and inter-protein contacts found in solved protein structures from the protein structure database PDB (protein data bank) (12) and their interactions annotated in iPFAM. A very similar approach has been taken by Stein et al. (13) resulting in the 3did database of domain contacts.
In addition to physical binding, individual domains can be linked by common biochemical or cellular functions. In the prediction of domain–domain interactions, we often cannot distinguish between physical and functional relations and treat physical binding as a special case of a functional link. The same is true for most techniques predicting protein–protein interactions.
Based on the well-known method of protein phylogenetic profiling, we had introduced the idea of domain phylogenetic profiling and demonstrated its utility for linking functionally related and physically interacting proteins (14). Other approaches to building domain interaction networks include the ‘domain team’ approach (15) that identifies functionally coupled domains based on their chromosomal location, as well as direct experimental evidence.
DIMA—the domain interaction map—was launched in 2005 as an online platform for our domain phylogenetic profiling approach and was soon extended to also include physical domain contacts from iPfam (16). In the greatly improved and extended 2.0 release, we have added new data sources and prediction methods: iPFAM domain contacts are now complemented by a similar dataset from 3did, domain profiling now covers the latest Pfam release and has more than doubled the number of genomes used for profiling to 460. With the domain pair exclusion algorithm (DPEA), we have integrated another prediction algorithm, which we apply to experimental and predicted data. Finally, we now use the SIMAP resource for highly efficient domain profiling. Below, we report in detail on the state of DIMA, which allows users to explore protein domain networks based on links produced by both experimental evidence and domain-relation predictions.
DOMAIN INTERACTIONS FROM KNOWN PROTEIN STRUCTURES
As for all areas of biology, experimental support of domain interactions is the most reliable source of data. While in the case of protein–protein interactions, a wealth of data has been collected in several well-maintained databases (17–23), the situation is quite different for domains. For the majority of protein–protein interactions found in the literature, no detailed information on the domains mediating the contact is provided and no comprehensive large-scale experiments for domain interactions are available to our knowledge.
We have included two datasets of domain contacts derived from solved protein structures in the PDB (12) into DIMA: iPfam, integrated with the well-known Pfam resource (11) and 3did (13) which represents an independent database with a similar scope at the EMBL.
Both of these datasets contain physical domain–domain interactions in separate protein chains as well as contacts within the same chain and are currently the only gold standard datasets available. Currently, iPfam and 3did contain 3074 and 3034 unique domain pairs, respectively. The union of both sets contains 3653 distinct domain pairs.
DOMAIN INTERACTIONS FROM COMPREHENSIVE PROTEIN INTERACTION DATA
As stated in the introduction, beyond direct physical binding the term protein interactions is often used to indicate functional coupling between proteins involved in the same signaling pathway or catalyzing subsequent steps of a biochemical reaction. The same is obviously true for conserved domains. To our knowledge, no databases of experimentally supported functional interactions among protein domains exist, beyond the protein structure based domains contacts described above.
Given the obvious incompleteness of the physical contact datasets and the non-existence of functional interaction data, prediction of domain relations is of great importance for our understanding of the role and contribution of modular proteins to the systems-level mechanisms of the cell.
An important approach to identifying interacting domains is built upon the idea that interacting domain pairs will be overrepresented in pairs of interacting proteins and this signal can be detected by statistical analysis. Many variations and improvements of the idea have been put forward (6–10). One of the best methods as of today is the DPEA by Riley et al. (6), which uses the expectation maximization algorithm to produce a maximum-likelihood estimate of the probability of interaction for domain pairs and evaluates the contribution of each pair of putative interacting domains by a modified likelihood ratio test (E-scores).
In order to get the best results possible, it is of great importance to run the algorithm on a large high-quality dataset of protein–protein interactions. With the recent formation of the international molecular exchange consortium (IMEx) and the resulting exchange of interaction data among the major players in the PPI field (DIP, IntAct, MINT, MPact, BioGRID and BIND), the task of obtaining a comprehensive dataset has become simple, as the archival resources DIP and IntAct will hold all relevant data. In DIMA, we use the PSIMItab data from IntAct (release date 2007-08-31) (22). The dataset provides 124 935 pairwise protein interactions from which we extracted 83 969 unique pairs from 159 different species for which Uniprot IDs were available. Pfam domain annotation for all proteins involved was obtained from the Uniprot-Swissprot and Uniprot-TREMBL data (24) resulting in a total of 126 260 possible interacting domain pairs.
DOMAIN PHYLOGENETIC PROFILING
Phylogenetic profiling was introduced as a means of predicting functional links and physical interactions among proteins by analyzing the presence or absence of orthologs over a large number of genomes (25). Proteins linked by common function were found to have correlated phylogenetic profiles—i.e. be either both present or absent from a given genome. The method has proven very useful for assigning functional annotation to novel proteins and newly sequenced organisms.
In DIMA, we apply the phylogenetic profiling approach to conserved domain represented by hits of the Pfam HMMs. As of version 2.0, DIMA domain profiling is carried out on 460 completely sequenced prokaryotic and eukaryotic genomes. Comprehensive Pfam domain coverage of all sequences was provided by SIMAP (26).
DOMAIN INTERACTIONS FROM PREDICTED PROTEIN INTERACTIONS
Despite great advances in coverage of the interactome of many important model organisms in recent years, the available data is still far from complete. Therefore, prediction of protein–protein interactions and functional relations is a very important addition. The STRING database (27) unites a large number of prediction approaches and experimental data resulting in a comprehensive scored list of predicted interactions. Although not as reliable as interactions supported by experimental evidence, the resource is reputed to produce high-quality results.
DIMA includes domain interaction predictions by the DPEA using the predicted interactions from STRING release 6.3 to complement the data derived from IntAct. Combined STRING scores were computed on the purely predicted evidence categories and a conservative threshold of 0.9 was applied to yield a set of high confidence PPI predictions for the subsequent DPEA analysis.
AVAILABILITY
DIMA is available at http://mips.gsf.de/genre/proj/dima. The web interface allows easy searching by domain identifier, domain description or sequence. Preferences such as phylogenetic profiling distance metrics and thresholds, entropy filtering, DPEA cutoffs and selection of organisms to be profiled can easily be changed by the user.
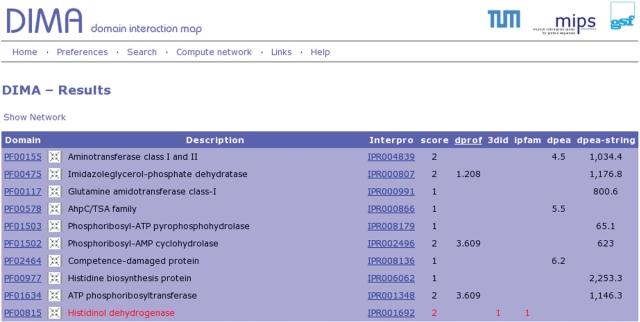
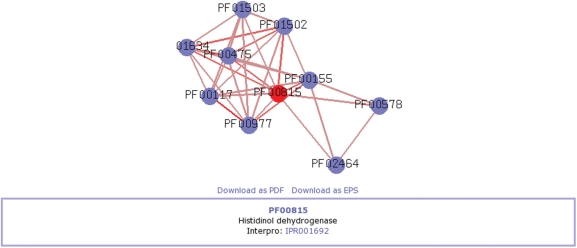
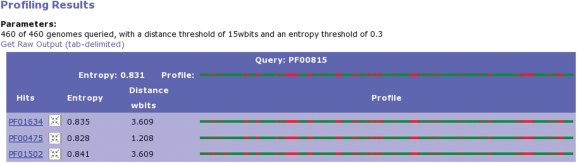
Results are primarily presented in a concise table format (Figure 1) showing the predictions and data sources supporting the domain relations and the user can choose to view a graphical representation of the local domain neighborhood (Figure 2) or details on the domain phylogenetic profiling results (Figure 3). The network can be navigated by centering any domain in the neighborhood and re-computing its respective interactions.
Figure 1.
Results are presented in a concise table showing all relevant information.
Figure 2.
If desired, a graphical representation of the local domain neighborhood is shown.
Figure 3.
For the profiling method detailed results can be examined or raw data downloaded.
For large-scale analysis or incorporation of DIMA data in own projects, we offer the option to compute the entire domain interaction network and return the results to the user by email upon completion. The DIMA backend program is available from the authors on request.
SYSTEM ARCHITECTURE
The pre-processing and backend software was written in Python with the exception of the domain profile neighbor search, which was implemented in C++ for performance reasons. The backend tool returns results as tab-separated tables for easy parsing and analysis outside the web environment. The web frontend was implemented in Java as part of the GenRe framework at MIPS and handles all user interactions.
CONCLUSION AND OUTLOOK
In addition to keeping DIMA up to date in terms of available data and user options, future work will comprise adding new algorithms and adding/updating relevant data sources as they become available. An important goal for the future is the derivation of a useful combined scoring scheme for all methods. Currently, this is hardly possible due to the extreme scarcity and methodological bias (exclusively protein structure data) of experimentally validated domain interaction data that are a prerequisite for a meaningful calibration.
Since its original publication, DIMA has evolved from a domain phylogenetic profiling platform to an integrated resource for domain interaction prediction. Of course, DIMA is not the only resource with respect to domain interactions. InterDom (28,29) is a service with similar goals but focuses more on explaining protein interactions with predicted domain interactions and to our knowledge incorporates fewer methods than DIMA. Using up-to-date data, a huge set of completely sequenced genomes and state-of-the-art algorithms DIMA provides a comprehensive resource of domain interactions with great value to the user.
ACKNOWLEDGEMENTS
A part of this work was funded by a grant from the German Federal Ministry of Education and Research (BMBF) within the BFAM framework (031U112C). The authors wish to thank Thomas Rattei and Roland Arnold for their help with the SIMAP resource and Philip Wong for careful reading of the manuscript. Funding to pay the Open Access publication charges for this article was provided by GSF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. Smart 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henikoff JG, Henikoff S, Pietrokovski S. New features of the blocks database servers. Nucleic Acids Res. 1999;27:226–228. doi: 10.1093/nar/27.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, et al. New developments in the interpro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 6.Riley R, Lee C, Sabatti C, Eisenberg D. Inferring protein domain interactions from databases of interacting proteins. Genome Biol. 2005;6:R89. doi: 10.1186/gb-2005-6-10-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng M, Mehta S, Sun F, Chen T. Inferring domain-domain interactions from protein-protein interactions. Genome Res. 2002;12:1540–1548. doi: 10.1101/gr.153002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C, Marcos F, Kanaan SP, Wuchty S, Chen DZ, Izaguirre JA. Predicting protein-protein interactions from protein domains using a set cover approach. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2007;4:78–87. doi: 10.1109/TCBB.2007.1001. [DOI] [PubMed] [Google Scholar]
- 9.Kim WK, Park J, Suh JK. Large scale statistical prediction of protein-protein interaction by potentially interacting domain (PID) pair. Genome Inform. Ser. Workshop Genome Inform. 2002;13:42–50. [PubMed] [Google Scholar]
- 10.Sprinzak E, Margalit H. Correlated sequence-signatures as markers of protein-protein interaction. J. Mol. Biol. 2001;311:681–692. doi: 10.1006/jmbi.2001.4920. [DOI] [PubMed] [Google Scholar]
- 11.Finn RD, Marshall M, Bateman A. iPfam: visualization of protein-protein interactions in PDB at domain and amino acid resolutions. Bioinformatics. 2005;21:410–412. doi: 10.1093/bioinformatics/bti011. [DOI] [PubMed] [Google Scholar]
- 12.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide protein data bank (wwpdb): ensuring a single, uniform archive of pdb data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein A, Russell RB, Aloy P. 3did: interacting protein domains of known three-dimensional structure. Nucleic Acids Res. 2005;33:D413–D417. doi: 10.1093/nar/gki037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagel P, Wong P, Frishman D. A domain interaction map based on phylogenetic profiling. J. Mol. Biol. 2004;344:1331–1346. doi: 10.1016/j.jmb.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Pasek S, Bergeron A, Risler JL, Louis A, Ollivier E, Raffinot M. Identification of genomic features using microsyntenies of domains: domain teams. Genome Res. 2005;15:867–874. doi: 10.1101/gr.3638405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagel P, Oesterheld M, Stümpflen V, Frishman D. The DIMA web resource – exploring the protein domain network. Bioinformatics. 2006;22:997–998. doi: 10.1093/bioinformatics/btl050. [DOI] [PubMed] [Google Scholar]
- 17.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. Mint: the molecular interaction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pagel P, Kovac S, Oesterheld M, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Mark P, Stümpflen V, et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 19.Güldener U, Münsterkötter M, Oesterheld M, Pagel P, Ruepp A, Mewes HW, Stümpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bader GD, Betel D, Hogue C.WV. BIND: the biomolecular interaction network database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xenarios I, Salwínski L, Duan XJ, Higney P, Kim SM, Eisenberg D. DIP, the database of interacting proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, et al. Intact – open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. Biogrid: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.UniProt Consortium. The universal protein resource (Uniprot) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rattei T, Arnold R, Tischler P, Lindner D, Stümpflen V, Mewes HW. Simap: the similarity matrix of proteins. Nucleic Acids Res. 2006;34:D252–D256. doi: 10.1093/nar/gkj106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Krüger B, Snel B, Bork P. String 7 – recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng SK, Zhang Z, Tan SH. Integrative approach for computationally inferring protein domain interactions. Bioinformatics. 2003;19:923–929. doi: 10.1093/bioinformatics/btg118. [DOI] [PubMed] [Google Scholar]
- 29.Ng SK, Zhang Z, Tan SH, Lin K. InterDom: a database of putative interacting protein domains for validating predicted protein interactions and complexes. Nucleic Acids Res. 2003;31:D251–D254. doi: 10.1093/nar/gkg079. [DOI] [PMC free article] [PubMed] [Google Scholar]