Abstract
The Plant Ontology Consortium (POC, http://www.plantontology.org) is a collaborative effort among model plant genome databases and plant researchers that aims to create, maintain and facilitate the use of a controlled vocabulary (ontology) for plants. The ontology allows users to ascribe attributes of plant structure (anatomy and morphology) and developmental stages to data types, such as genes and phenotypes, to provide a semantic framework to make meaningful cross-species and database comparisons. The POC builds upon groundbreaking work by the Gene Ontology Consortium (GOC) by adopting and extending the GOC's principles, existing software and database structure. Over the past year, POC has added hundreds of ontology terms to associate with thousands of genes and gene products from Arabidopsis, rice and maize, which are available through a newly updated web-based browser (http://www.plantontology.org/amigo/go.cgi) for viewing, searching and querying. The Consortium has also implemented new functionalities to facilitate the application of PO in genomic research and updated the website to keep the contents current. In this report, we present a brief description of resources available from the website, changes to the interfaces, data updates, community activities and future enhancement.
INTRODUCTION
The increasing availability of data from several plant genome sequencing projects provides a promising direction for investigating genes and their functional and sequence homologs involved in plant development. Historically, the data that could be used in comparative studies would often reside in different, species-specific databases, each using slightly different terminology, creating a barrier for researchers trying to answer cross-species questions relating to phenotypes. A solution to this data integration challenge is the creation of a controlled vocabulary of common, consistent and internationally recognized descriptive terminology that can be shared and used uniformly among species and to which genetic and phenotypic data can be associated.
The Plant Ontology (PO) is a community resource designed to fulfill the need for uniform terminology to describe plant structure and developmental stages (1), it was initially developed (1–3) based on ontologies for the plant model species, Arabidopsis, rice and maize, developed by The Arabidopsis Information Resource [TAIR; (4)], Gramene (5,6), and MaizeGDB (7), respectively. Terms are continuously added as needed to describe other angiosperms such as Triticeae (wheat, oat and barley), Solanaceae (tomato, potato and peppers), Fabaceae (soybean and Medicago), Rutaceae (citrus), Asteraceae/Compositae (sunflower and lettuce) and Salicaceae (poplar, a tree species), to enable comparative plant genomics research. The PO is included in the open-source Open Biomedical Ontologies (OBO) project hosted at http://obofoundry.org, a resource for sharing controlled vocabularies for different biological domains.
POC WEBSITE UPDATES
Highlights of POC homepage
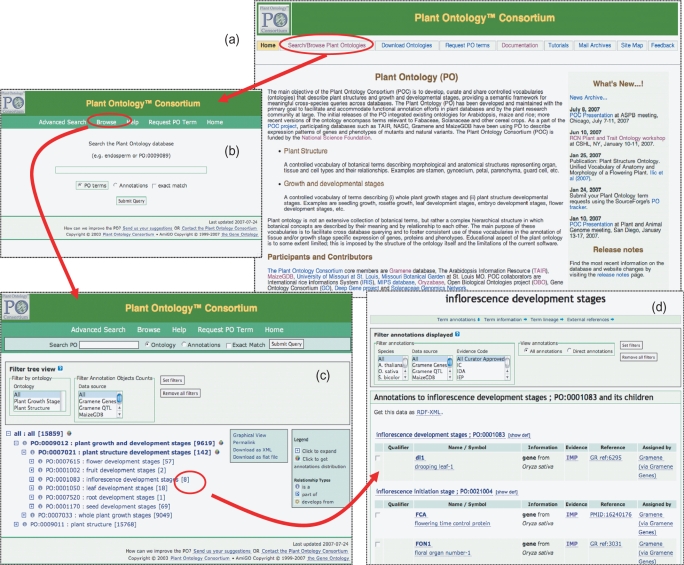
The POC website http://www.plantontology.org was publicly launched in 2004 (Figure 1a). The front page serves as an entry point for users to browse recent POC activities and displays links to the ontology browser for PO and available annotations, downloadable ontology and annotation files, documentation and tutorials. It is designed as an interactive environment to facilitate community participation in the development and utilization of the ontologies where users are encouraged to send comments through ‘Feedback’ link to the developers or by subscribing the POC mailing lists to participate in discussions and get announcements. We recently added a new feature ‘Request PO terms’ on the navigation menu that directs users to the ‘PO TERM requests’ tracker in SourceForge (http://sourceforge.net/tracker/?group_id=76834&atid=835555) and allows PO editors and users to request new PO terms or changes to existing terms. The tracker streamlines the monitoring process of adding and changing the ontology terms and encourages community participation.
Figure 1.
Stepwise guide to search and browse the PO and annotations. (a) The view of Plant Ontology project website (http://www.plantontology.org). Display features a navigation menu, ‘what's new’ section and the project description. The ‘Search/Browse Plant Ontologies’ links to the front page of PO ontology browser. (b) The simple search page for Ontology database with several options. All ontology browser pages have the same header, which allows users to search for PO terms or annotations, request PO term if it is not available in the database and provides direct access to a ‘Help’ page that describes details of how to use the ontology browser. The ‘Advanced Search’ link allows users to search for more than one term or annotations in different fields of the database and the search results can be filtered by ontology type, the evidence code, source species and data source. The ‘Browse’ link opens up a browsable ontology tree. (c) Browsable tree view of ontology. It can be browsed by expanding (click on + sign) or contracting (click on − sign) on the left side of the term name. The circular icons with characters I/P/D to the left of the term represent the relationship types, IS_A, PART_OF and DEVELOPS_FROM, respectively. More options on pie chart of annotations, links to download in XML or GO flat file formats, Permalink for book marking and summary of the number of annotations next to term names is provided. To view the annotations e.g. associated with PO:0001083: inflorescence development stages, click the number ‘8’ in curved brackets. (d) A list of annotations associated to a term (e.g. PO:0001083). The annotation table provides information on object type (gene, QTL, stock), name of the object, synonyms, link to the same entry in source database, evidence and evidence code used for associating the ontology term to the object.
New look of PO browser
The major change in the POC website since its debut 3 years ago is the new interface of the PO browser (http://www.plantontology.org/amigo/go.cgi; Figure 1b). The PO browser is a modified version of the AmiGO browser originally developed by the Gene Ontology Consortium (GOC) (8); it is an Internet-based application written in Perl that enables users to browse and query data from any OBO Foundry-compatible database and to view the ontology as a graph structure (Figure 1c). In addition to viewing the terms and definitions, the browser also provides a view for annotations associated to a term (Figure 1d). Initially the source code for the GOC browser allowed the display of only one type of annotation object, ‘gene product’. However, the PO project required annotation for multiple object types, including genes, germplasms and quantitative trait loci (QTLs), therefore we customized the browser and contributed our modifications to the AmiGO source project, which is available for download at the GO website (http://www.godatabase.org/dev/). We are regularly updating and integrating improvements in the browser by maintaining the most recent version of AmiGO. A simple case of using new PO browser is illustrated in the Supplementary Data (Supplementary Figure S1).
ONTOLOGY UPDATES
The POC project was initiated in 2003 (Figure 1a), and in the first 3 years POC released two major ontologies: (i) Plant Structure Ontology (PSO), which, as one of the top-level parent terms in PO, describes morphological and anatomical structures and includes organs and organ systems, tissues and cell types (2); and (ii) whole plant Growth Stages Ontology (GSO), which describes organism growth stages such as ‘germination’, ‘rosette growth’, ‘flowering’ and ‘senescence’ that cover the vegetative and reproductive lifecycle of an entire plant (3). The ontology development and applications are described in detail by Ilic et al. (2) and Pujar et al. (3), respectively.
In addition to the PSO and GSO published earlier, we report for the first time the addition of the Plant Structure Development Stages Ontology (PSDSO), which is designed to describe developmental stages of individual plant structures of a generic flowering plant, namely the flower, leaf, fruit, inflorescence, root and seed. Instead of providing detailed species-specific staging in a form of a ‘parallel bins’ for each structure, we opted to integrate existing species-specific plant structure developmental stages defined in the temporal ontologies provided for Arabidopsis by TAIR, maize by MaizeGDB and rice by Gramene and Oryzabase databases [Oryzabase; (9)], thus creating a standardized staging system for each plant organ and organ system. This ontology is the first multi-species representation of developmental stages of plant structural parts in angiosperms. Although derived from staging systems developed for Arabidopsis, maize and rice, it is designed to accommodate subsequent incorporation of developmental stages of several other angiosperms, such as those representing Solanaceae and Fabaceae taxonomy families. Currently 123 terms are arranged in six main nodes in the PSDSO (Figure 1c). As of the June 2007 release of the POC database, there are 142 genes and gene products annotated to it and its children. Figure 1d shows the annotations to PO:0001083, inflorescence development stages and its children. Together with GSO, the PSDSO is placed as a subset of a grouping term ‘Plant Growth and Development Stages PO:0009012’, which becomes one of the top-level parent terms of the PO (Figure 1c) and carries its own namespace in formal ontology design and practices suggested by OBO Foundry.
PO is cross-referenced to cell ontology
Cell ontology (CL) was developed to describe cell types that cover prokaryotic, fungal, animal and plant kingdoms (10). Many CL terms under the Plant Cell node are identical to terms in the PO. In such cases, the corresponding CL identifier is added as cross reference to the PO term and is listed in ‘External References’ of ‘Term Details’ page.
Newly added annotations and PO applications
In 2004, with the first release of the PSO and GSO, associations were available for more than 3400 gene annotations from Arabidopsis, rice and maize. These associations were contributed by three species-specific databases: TAIR, Gramene and MaizeGDB. Within 3 years, the number of annotations and the data types has increased from about 3400 to 32 000 for approximately 16 000 unique genes, QTLs and germplasms. The recent additions include annotations to 1897 germplasms by Nottingham Arabidopsis Stock Centre (NASC, http://arabidopsis.info) in 2006; more than 8000 rice QTLs contributed by Gramene in March 2007; and 6241 genes assigned by TAIR in July 2007. These statistics are updated in each database release and can be found in ‘Release notes’ section (Figure 1a).
In the past year, several databases have incorporated PO to associate with object types for their research interests, which are listed in Table 1. For example, the International Rice Information System [IRIS; (11)] is using PO to describe and catalog their mutant phenotypes. Phenotypes for plants are codified using terms for plant anatomy and developmental stages. Researchers from the International Rice Research Institute (IRRI; http://www.irri.org) and their collaborators from Bioversity International (http://www.bioversityinternational.org) are collaborating with the POC to systematically index descriptions for interesting phenotypes relating to agronomic traits, such as yield, biotic and abiotic stress tolerance, as well as improved grain quality. In the future, we anticipate new annotations from existing collaborators and additional community contribution to come from functional genomics projects and other model organism databases, such as the Solanaceae Genomic Network [SGN; (12)].
Table 1.
List of some biological databases that have incorporated PO to describe and catalog their object types
| Database name and website | Materials annotated to PO |
|---|---|
| Bioassay and Phenotype Database (BAP DB) http://bioweb.ucr.edu/bapdb/ | Mutants, genes |
| BRENDA (The Comprehensive Enzyme Information System) http://www.brenda.uni-koeln.de/ | Enzymes |
| Gramene (A Resource for Comparative Grass Genomics) http://www.gramene.org | Genes, QTLs, proteins |
| Genevestigator (Microarray Database and Analysis Toolbox) http://www.genevestigator.ethz.ch/ | Microarray |
| IRIS (International Rice Information System) http://www.iris.irri.org | Mutants |
| MaizeGDB (Maize Genetics and Genomics Database) http://www.maizegdb.org | Genes, Mutants |
| NASC (Nottingham Arabidopsis Stock Center) http://arabidopsis.info/ | Germplasms |
| Oryza Tag Line http://urgi.versailles.inra.fr/OryzaTagLine/ | Mutants |
| PLEXdb (Plant Expression Database) http://www.plexdb.org | Microarray data |
| Rice Oligonucleotide Array Project http://www.ricearray.org | Microarray data |
| SGN (Solanaceae Genomics Network) http://www.sgn.cornell.edu | Genes |
| TAIR (The Arabidopsis Information Resource) http://www.arabidopsis.org | Genes, germplasms, microarray data |
| TRIM (Taiwan Rice Insertion Mutants Database) http://trim.sinica.edu.tw/ | Mutants |
In order to provide users with a quick way to find functional information of organ- or tissue-specific genes in their expression, the PO browser provides an external link to the GO gene product detail page (Supplementary Figure S1). This link takes the users to the GO database, an external site, where they will be able to view the protein sequence, perform BLAST queries and find the genes’ functional characterization (roles played in biological process, localized in a cellular component(s) and the molecular functions) that are predominant in the selected plant tissue or organ. Currently this feature is for Arabidopsis genes only; we are working with the GOC group and other plant model organism databases to bring more GO associations from other plant species to PO database. We anticipate that linking to GO will facilitate the application of PO in plant genomics research.
A ‘Help’ page describing how to use the PO ontology browser in more detail is available at http://www.plantontology.org/amigo_user_guide/index.html or by clicking on the ‘Help’ link in the navigation bar at the top of all browser pages. Users are expected to find more details about genotype–phenotype association(s) in the original source database. An example with gene to QTL phenotypes associations found in the Gramene database is provided in the Supplementary Data (Supplementary Figure S2).
DATA SOURCES AND DATA AVAILABIITY
All information described on the website is freely available. The POC CVS (concurrent versioning system) repository is used to maintain the ontology as flat files that describe terms, relationships and definitions. These are kept in both OBO and GO formats (http://www.geneontology.org/GO.format.shtml—flatfile) and described in the ‘Documentation’ section of the website. The ontology flat files namely the po_anatomy.obo (PSO) and po_temporal.obo (GSO, PSDSO) can be downloaded for local use or to build annotations by visiting the ‘Download Ontologies’ section of the website. It provides instructions on how gain anonymous access to POC CVS repository using the command line interface or from the webCVS viewer. Species-specific ontologies from TAIR, Gramene and MaizeGDB are also available in this section. The MySQL database dumps of the most current and previous versions of the ontology database are available also freely.
Ontology flat files in OBO format are also maintained in the OBO SourceForge CVS repository (http://obofoundry.org) and are updated daily from the POC CVS repository, which contains the most up-to-date version of the files.
Association files that contain the annotations from contributing databases and projects constitute the primary source used by the POC database to relate objects such as gene, germplasm (stock/cultivar/mutant) and QTL from individual species to ontology terms. The association data includes the name of the database that provided the annotation, database object ID, symbol of the object, PO accession ID, the object's full name, object type, species, synonyms and the evidence that was used to make the annotation. These files use a standardized tab-delimited format originally developed by the GOC that is convenient to read and parse. More information on the association file format is found at http://www.plantontology.org/docs/otherdocs/assoc-file-format.html. Evidence codes are described at http://www.plantontology.org/docs/otherdocs/evidence_codes.html. Projects wishing to contribute annotations may communicate with the POC to submit the file datasets. Regular contributors are given the permission to commit the file directly to the POC CVS repository. Quality assurance of the annotated data provided on the project site is the sole responsibility of contributing databases.
The most current versions of the ontology, associations and mapping flat files from the POC CVS are used to populate the Plant Ontology MySQL database using the GO-dev toolkit, downloaded from http://www.godatabase.org/dev/. Tools and scripts were either downloaded from the GO-dev toolkit site or developed in-house to automate the process, refresh the POC CVS, generate the database schema, load data from the different file formats and make MySQL database dumps available for local use.
Mapping PO to vocabularies from external sources
Before PO was developed and recognized by the plant research community, several plant species databases, such as Gramene, MaizeGDB and TAIR, had already created their own controlled vocabularies to describe rice, maize, Triticeae and Arabidopsis developmental and growth stages. Most of these vocabularies are retired now and they use the PO. To help external database users adopt PO in their projects during the transition period, the suggested mapping files are generated and used to convert species-specific vocabularies to PO or to map users’ annotations to PO. These files are available via CVS (http://brebiou.cshl.edu/viewcvs/Poc/mapping2po/), which contains database cross-references that link PO terms to the terms in external databases that maintain specialized or species-specific vocabulary. Database abbreviations used in ‘External References’ found in ‘Term Details’ page are described in http://www.plantontology.org/docs/dbxref/PO_DBXref.txt. More information on mapping file usage and format may be accessed online (http://www.geneontology.org/GO.format.shtml#mappings). We encourage existing plant species-specific databases that are not currently using PO in their datasets to follow this approach to contribute their species-specific ontology and the annotations to the POC database.
OUTREACH ACTIVITIES
The consortium was initiated with a goal to ‘allow the fruits of research in one plant species to be more easily used in the study of other species, leading to a greater understanding of plant biology’; this continues to be the driving force of the consortium. To achieve this, POC members have actively participated in a variety of conferences in past three years through poster presentations, oral presentations and workshops. Details of these outreach activities are available from the ‘Documentation’ link available in the website navigation bar. These workshops include users’ group meetings to work with ontology developers, computational scientists and database curators to discuss and review PO for better use. Community-engaging efforts are ongoing and will continue to be a high priority for the project to facilitate the improvement and utility of the ontology for plant researchers.
FUTURE ENHANCEMENTS
The POC is looking forward to integrate species-specific terms for legumes (soybean and Medicago), Solanaceae, wheat and poplar. We expect further contribution of annotations from participating databases and external collaborating groups while introducing new annotation types. We plan to continue to enhance the project website through software updates and additional datasets. New functionalities to the ontology browser, such as displaying phenotypic description, a ‘slim’ version of the ontology (PO terms with most common usage), new search filters (e.g. by annotation object types) and integrating GO associations from other plant species in POC database are being developed. As resources allow, we hope to add species-specific anatomical and developmental landmark images that are annotated to PO terms, together with an image ontology (image.obo in http://obofoundry.org), and extend POC to serve as an educational resource for the plant community.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
ACKNOWLEDGEMENTS
The Plant Ontology Consortium gratefully acknowledges the Gene Ontology Consortium for the contributed infrastructure, software and kind support. Special thanks to Chris Mungall and John Day-Richter from the GOC, to Peter Van-Buren, our system administrator at Cold Spring Harbor Laboratory and Ken Youens-Clark for critical review of the manuscript. We also thank all database groups, annotation contributors, researchers, curators, reviewers, collaborators, specifically Alice Clare Augustine from Monsanto and David Selinger and Arthur Lane from Pioneer Hi-Bred International companies, and users who have participated in the effort. Full list of collaborators and contributors is available at http://www.plantontology.org/docs/otherdocs/collab.html and http://www.plantontology.org/docs/otherdocs/acknowledgment_list.html. This project is supported by the National Science Foundation (Grant No. 0321666) to the Plant Ontology Consortium and the U.S. Department of Agriculture-Agricultural Research Service. Funding to pay the Open Access publication charges for this article was provided by National Science Foundation (Grant No. 0321666).
Conflict of interest statement. None declared.
REFERENCES
- 1.Jaiswal P, Avraham S, Ilic K, Kellogg E, McCouch S, Pujar A, Reiser L, Seung RY, Sachs MM, Schaeffer M, et al. Plant Ontology (PO): a controlled vocabulary of plant structures and growth stages. Comp. Funct. Genomics. 2005;6:388–397. doi: 10.1002/cfg.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ilic K, Kellogg E, Jaiswal P, Zapata F, Stevens PF, Vincent LP, Avraham S, Reiser L, Pujar A, et al. Plant Structure Ontology. Unified vocabulary of anatomy and morphology of a flowering plant. Plant Physiol. 2007;143:587–599. doi: 10.1104/pp.106.092825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pujar A, Jaiswal P, Kellogg E, Ilic K, Vincent L, Avraham S, Stevens P, Zapata F, Reiser L, et al. Whole Plant Growth Stage Ontology for angiosperms and its application in plant biology. Plant Physiol. 2006;142:414–428. doi: 10.1104/pp.106.085720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berardini T, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, et al. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 2004;135:745–755. doi: 10.1104/pp.104.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaiswal P, Ni J, Yap I, Ware D, Spooner W, Youens-Clark K, Ren L, Liang C, Zhao W, et al. Gramene, a bird's eye view of cereal genomes. Nucleic Acids Res. 2006;34:D717–D723. doi: 10.1093/nar/gkj154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamazaki Y, Jaiswal P. Biomedical ontologies in rice databases. An introduction to the activities in Gramene and Oryzabase. Plant Cell Physiol. 2005;46:63–68. doi: 10.1093/pcp/pci505. [DOI] [PubMed] [Google Scholar]
- 7.Vincent PL, Coe EH, Polacco M. Zea mays ontology - a database of international terms. Trends Plant Sci. 2003;8:517–520. doi: 10.1016/j.tplants.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 8.Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurata N, Yamazaki Y. Oryzabase. An integrated biological and genome information database for rice. Plant Physiol. 2006;140:12–17. doi: 10.1104/pp.105.063008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bard JB, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLaren CG, Bruskiewich RM, Portugal AM, Cosico AB. The International Rice Information System. A platform for meta-analysis of rice crop data. Plant Physiol. 2005;139:637–642. doi: 10.1104/pp.105.063438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, et al. The SOL Genomic Network. A comparative resource for Solanaceae biology and beyond. Plant Physiol. 2005;138:1310–1317. doi: 10.1104/pp.105.060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.