Abstract
The Zebrafish Information Network (ZFIN, http://zfin.org), the model organism database for zebrafish, provides the central location for curated zebrafish genetic, genomic and developmental data. Extensive data integration of mutant phenotypes, genes, expression patterns, sequences, genetic markers, morpholinos, map positions, publications and community resources facilitates the use of the zebrafish as a model for studying gene function, development, behavior and disease. Access to ZFIN data is provided via web-based query forms and through bulk data files. ZFIN is the definitive source for zebrafish gene and allele nomenclature, the zebrafish anatomical ontology (AO) and for zebrafish gene ontology (GO) annotations. ZFIN plays an active role in the development of cross-species ontologies such as the phenotypic quality ontology (PATO) and the gene ontology (GO). Recent enhancements to ZFIN include (i) a new home page and navigation bar, (ii) expanded support for genotypes and phenotypes, (iii) comprehensive phenotype annotations based on anatomical, phenotypic quality and gene ontologies, (iv) a BLAST server tightly integrated with the ZFIN database via ZFIN-specific datasets, (v) a global site search and (vi) help with hands-on resources.
INTRODUCTION
The zebrafish is an established model for studies in vertebrate biology. Significant contributions are made to the identification and characterization of genes and pathways involved in development, organ function, behavior and disease. As the zebrafish model organism database, ZFIN facilitates research by providing a central database and a uniform interface to integrate and view the large amount of diverse data (1). ZFIN data derive from expert manual curation of scientific literature, collaboration with major resource centers and submissions from individual investigators. Curation is an ongoing effort with daily updates and enhancements. All data are attributed to their primary source. Annotations using controlled vocabulary terms from biomedical ontologies are a part of our routine curation efforts. These annotations provide a standardized data representation that enables cross-species comparisons. Extensive links to outside resources from ZFIN data pages further enhance cross-species comparisons. Data curated at ZFIN are also available through centers such as NCBI/Entrez Gene (2), Vertebrate Genome Annotations (Vega) (3) and the Ensembl (4) and UCSC (5) genome browsers.
A NEW LOOK AT ZFIN
ZFIN's new home page serves as a hub for online zebrafish resources. Major enhancements include: improved grouping and prioritization of ZFIN's most popular resources based on community input and page usage statistics; prominent access to the Zebrafish International Resource Center (ZIRC); expanded genome resource links and a ‘News’ section. The three-tab navigation bar available on all ZFIN pages provides convenient traversal of ZFIN and ZIRC pages.
ZFIN welcomes contributions to our ‘News’ section. Please email to: zfinadmn@zfin.org with your submissions.
EXPANDED SUPPORT FOR GENOTYPES AND PHENOTYPES
Support for mutant and transgenic fish has been expanded to include complex genotypes containing multiple mutations, zygosity and transgenic constructs. Major enhancements include:
Mutants/Transgenics query interface
This query interface (http://zfin.org/cgi-bin/webdriver?MIval=aa-fishselect.apg&line_type=mutant) provides the most comprehensive access to genotype and phenotype data. Mutant/Transgenic queries may include gene or allele name, chromosome, mutagen, mutation type and anatomical structures that are affected. Queries may be performed on both mutants and transgenics or constrained to either characterized mutants or transgenics. The query results page provides summary data and relevant links including allele name, parental zygosity, mutation type, affected gene, mapping position and curated phenotypes.
Genotype page
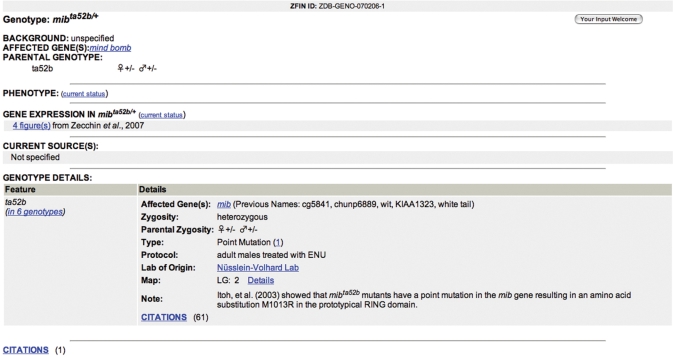
This new page is the hub for genotype-specific data for the more than 5000 genotypes described in ZFIN (Figure 1). Genetic background, parental genotypes, affected genes and current sources for fish with this genotype are listed. Links to all phenotype and gene expression data associated with the genotype are included. A ‘Genotype Details’ section lists affected genes, zygosity, parental zygosity, mutation type, mutagen protocol, lab of origin and mapping information for each allele. Links to construct pages are provided for transgenic genotypes. Mutant/Transgenics query results, phenotype and gene expression details pages and gene pages link to the genotype page.
Figure 1.
The genotype page summarizes genotype specific data for mibta52b/ta52b and provides extensive cross-linking to all mibta52b/ta52b data in ZFIN.
Construct page
This new page provides information about transgenic constructs including regulatory regions, coding sequences, associated lines and related publications.
Gene page
A ‘Mutants and Targeted Knockdowns’ section has been added to the gene page. This section contains links to all associated genotypes and phenotypes. Anatomical ontology (AO) (1) and gene ontology (GO)(6) terms describing the phenotype are summarized. Knockdown reagents are listed.
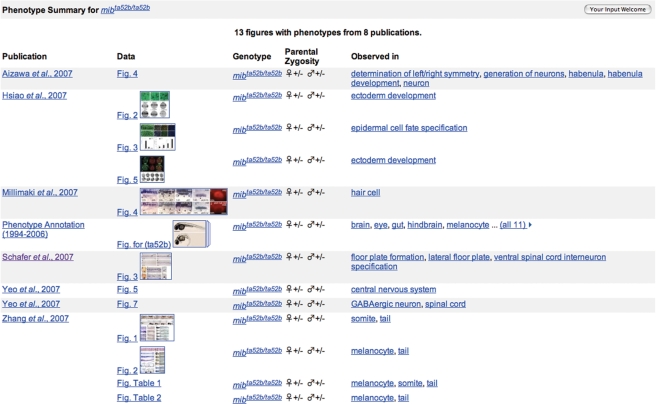
Phenotype summary page
Thumbnail images, genotype details, parental zygosity, supporting publications and links to affected structures and processes that characterize the phenotype can be accessed from this new page (Figure 2). Detailed phenotype annotations can be viewed by following the thumbnail or figure links. This page can be accessed from gene, morpholino and genotype pages.
Figure 2.
Phenotype summary page for mibta52b/ta52b. Publication, genotype and ‘observed in’ link to their respective pages. Figures and thumbnail images link to curated phenotype details.
Anatomy page
Links are available from anatomy pages to mutant and transgenic genotypes with related phenotypes.
Nomenclature
A consistent and informative system for naming genotypes and transgenic constructs has been developed in consultation with the Zebrafish Nomenclature Committee. These conventions are described at: http://zfin.org/zf_info/nomen.html.
COMPREHENSIVE PHENOTYPE ANNOTATIONS
Phenotype data from mutant screens and morpholino studies make the zebrafish a powerful model for elucidating gene function in development and disease. The correlation of zebrafish phenotype data with human genes and genetic syndromes is a major goal of zebrafish research. Our approach will facilitate cross-species comparisons by providing a comprehensive and consistent framework for phenotype annotation and queries.
Ontologies, hierarchical controlled vocabularies, provide the structure required for consistent annotations and subsequent comparisons. Ontologically derived annotations are successfully used by model organism databases to enhance computational and manual data analyses. GO annotations are an integral part of ZFIN's gene page and contribute to cross-species comparisons at AmiGO, the official GO query tool provided by the gene ontology consortium. AO annotations facilitate queries of gene expression data (http://zfin.org/cgi-bin/webdriver?MIval=aa-xpatselect.apg).
The AO and GO annotations can also be used for the robust classification of phenotypes, together with a cross-species structured vocabulary for describing phenotypes. The phenotypic quality ontology (PATO, http://www.bioontology.org/wiki/index.php/PATO:Main_Page) is being developed in collaboration with the model organism community and the National Center for Biomedical Ontology (NCBO)(http://www.bioontology.org/).
Phenotype annotations are based on a bipartite syntax composed of an entity and a quality that may be used to describe an observable structure or process. The entity, a GO or AO term, represents the part of the phenotype being described. The quality, a PATO term, characterizes how the entity is affected. Examples of PATO qualities are ectopic, fused, small, edematous and arrested. Special modifiers, or tags, are included in PATO to distinguish normal from abnormal phenotypes.
We now capture detailed phenotype data from the scientific literature as part of our daily curation. Our first 7 months of phenotype curation using PATO yielded nearly 5000 annotations from ∼200 publications. Phenotype descriptions cataloged at ZFIN in previous years and ongoing collaborations with laboratories conducting large-scale mutant screens provided an additional 9600 phenotype annotations for ∼3000 genotypes.
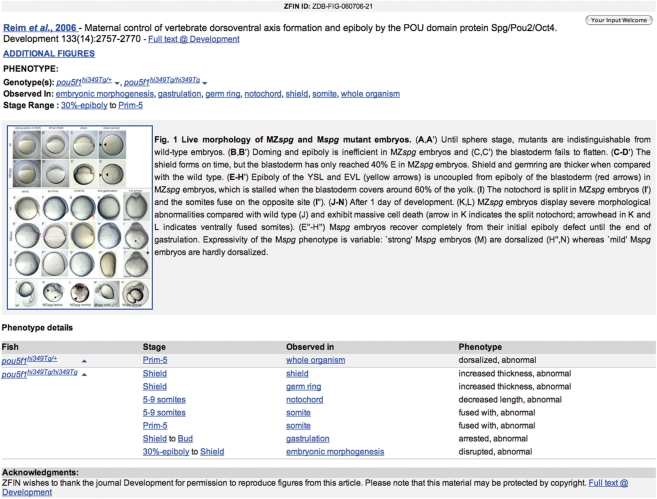
Phenotypes are tightly integrated into many parts of ZFIN and can be found on gene, genotype and anatomy pages. Comparisons of curated phenotypes at ZFIN are supported through the specification of ‘affected anatomy’ on the Mutants/Transgenics query form (http://zfin.org/cgi-bin/webdriver?MIval=aa-fishselect.apg&line_type=mutant). Phenotype annotations are associated with individual published figures and displayed on phenotype figure pages (Figure 3). Images and figure captions are included when consistent with journal copyright permissions. Ultimately, the use of PATO and other ontologies will provide a means for more complex queries at ZFIN and for comparisons with other organisms, specifically for the determination of animal models of human disease.
Figure 3.
A typical published figure with curated phenotype annotations. ‘Whole organism’, ‘shield’, ‘germ ring’, ‘notochord’ and ‘somite’ are entities derived from the zebrafish AO. ‘Gastrulation’ and ‘embryonic morphogenesis’ are entities derived from the GO. ‘Increased thickness’, ‘decreased length’, ‘malformed’, ‘fused with’, ‘arrested’, ‘disrupted’ and ‘abnormal’ are PATO qualities. Links are provided to the genotype page, stage description, anatomy term pages, GO details page and the full text of the publication. Figure reproduced from Reim and Brand (11).
We encourage the use of PATO by laboratories annotating phenotype data and provide tools for this purpose. Please email us at: zfinadmn@zfin.org for additional information.
ZFIN BLAST
Basic Local Alignment Search Tool, BLAST, is a powerful tool for performing comparisons of primary biological sequence data. ZFIN BLAST (http://zfin.org/cgi-bin/webdriver?MIval=aa-blast.apg) tightly integrates with the ZFIN database via ZFIN-specific data sets. Sequences or sequence accession identifiers (IDs) can be used to identify zebrafish genes annotated with phenotype, expression or GO data. Available data sets include, but are not limited to, ZFIN GenBank sequences, ZFIN cDNA sequences, ZFIN genes with expression, ZFIN morpholino sequences, ZFIN microRNA sequences and ZFIN Vega transcripts. ZFIN BLAST displays sequence alignment results with direct links to ZFIN gene and clone pages. Genes with associated expression, phenotype and gene ontology data are annotated with E, P and G icons, respectively. A camera icon indicates that representative figures are available at ZFIN.
ZFIN BLAST utilizes the WU-BLAST program (http://blast.wustl.edu/). Because multiple BLAST searches can require significant system resources, some reasonable constraints have been implemented to optimize overall performance. The sequence query length is limited to 50 000 letters. A graphical display is available for the first 50 alignments only. The zebrafish trace archive may be searched by single queries. Small to medium datasets accommodate batch queries of up to 100 sequences.
SITE SEARCH
The entire ZFIN site can be searched quickly using the ‘site search’ feature located at the top right of all ZFIN pages. A results table narrows the search by categorizing matching records by data type. The number of matching pages is displayed for each category. Links to exact matches are provided when available. All individual matching records are listed. This broad view of ZFIN data complements the more comprehensive data type-specific query forms.
HELP WITH HANDS-ON RESOURCES
ZFIN plays an important role in the identification and procurement of probes, clones and fish lines used in research. Clones, probes and knockdown reagents are listed on gene pages. Fish lines are found on genotype pages. Sources and ‘Order this’ links are provided to resource centers and laboratories that can provide the resource.
ZFIN works closely with ZIRC to create reciprocal links between ZFIN data pages and ZIRC inventory pages. Recent changes to ZFIN's home page and a new tab navigation system provide easy access to ZIRC.
ZFIN anatomy pages provide researchers with an easy means for identifying probes for anatomical structures. Probes, used in large-scale in situ hybridization screens (7–9), are ranked by intensity, specificity and contrast using a five-star system.
FUTURE DIRECTIONS
We will soon expand ZFIN to incorporate detailed information on antibodies that recognize zebrafish anatomical structures and gene products. Sequence support will be enhanced to include transcripts and repeats. Expression patterns and phenotypes will be associated with specific transcripts and transcript annotations. GBrowse, the generic genome browser provided by GMOD (10), will provide an interactive view of the zebrafish genome integrated with ZFIN's rich biological data. Expanded use of ontologies will accommodate more robust queries of ZFIN data.
IMPLEMENTATION
ZFIN is currently implemented with the IBM/Informix relational database management software. Web-based HTML forms combined with Java/JSP, JavaScript, Perl and CGI scripts provide access to the database. A detailed description of the current ZFIN data model can be found at http://zfin.org/DataModel.
ACKNOWLEDGEMENTS
Funds for the development of the Zebrafish Information Network are provided by the NIH HG002659. Funding to pay the Open Access publication charges for this article was provided by NIH HG002659. Funds to ZFIN through the NCBO to further development of tools and curation of phenotype annotation are provided by the NIH HG004028.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sprague J, Bayraktaroglu L, Clements D, Conlin T, Fashena D, Frazer K, Haendel M, Howe DG, Mani P, et al. The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashurst L, Chen C.-K, Gilbert JGR, Jekosch K, Keenan S, Meidl P, Searle SM, Stalker J, Storey R, et al. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2005;33:D459–D465. doi: 10.1093/nar/gki135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birney E, Andrews TD, Bevan P, Caccamo M, Chen Y, Clarke L, Coates G, Cuff J, Curwen V, et al. An overview of Ensembl. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, et al. The UCSC Genome Browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thisse C, Thisse B. High throughput expression analysis of ZF-models consortium clones. 2005. ZFIN Direct Data Submission ( http://zfin.org).
- 8.Thisse B, Thisse C. Fast Release Clones: a high throughput expression analysis. 2004. ZFIN Direct Data Submission ( http://zfin.org).
- 9.Thisse B, Pflumio S, Furthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, et al. Expression of the zebrafish genome during embryogenesis (NIH R01 RR15402). 2001. ZFIN Direct Data Submission ( http://zfin.org).
- 10.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;10:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reim G, Brand M. Maternal control of vertebrate dorsoventral axis formation and epiboly by the POU domain protein Spg/Pou2/Oct4. Development. 2006;133:2757–2770. doi: 10.1242/dev.02391. [DOI] [PubMed] [Google Scholar]