Abstract
The Shanghai RAPESEED Database (RAPESEED, http://rapeseed.plantsignal.cn/) was created to provide the solid platform for functional genomics studies of oilseed crops with the emphasis on seed development and fatty acid metabolism. The RAPESEED includes the resource of 8462 unique ESTs, of which 3526 clones are with full length cDNA; the expression profiles of 8095 genes and the Serial Analysis of Gene Expression (SAGE, 23 895 unique tags) and tag-to-gene data during seed development. In addition, a total of ∼14 700 M3 mutant populations were generated by ethylmethanesulfonate (EMS) mutagenesis and related seed quality information was determined using the Foss NIR System. Further, the TILLING (Targeting Induced Local Lesions IN Genomes) platform was established based on the generated EMS mutant population. The relevant information was collected in RAPESEED database, which can be searched through keywords, nucleotide or protein sequences, or seed quality parameters, and downloaded.
INTRODUCTION
Brassica species, including Brassica napus, B. oleracea and B. rapa, are important vegetable and oilseed crops. Seed of B. napus contains abundant fatty acids such as oleic acid and linoleic acid, and serves as one of the main sources of plant oil for human daily life. As the second oilseed crop in the world, compared to soybean, numerous studies have been performed on B. napus with the focus on yield increase and quality improvement through genetic breeding (1–3), and development of genetics and molecular biology tools has significantly contributed to the relevant studies. However, the deficiency of genome sequences and the detailed genomic information counteract the functional genomics studies of rapeseed, especially the genetics approach through T-DNA tagged mutant, of which the gene sequences and expression information are required (4–6). Thus, the increased information of gene sequences and expression profiles will be beneficial for rapeseed functional genomics studies and further bioengineering.
Currently, several databases for Brassica species are available. Handa reported a database functions on B. napus mitochondrial genome and the comparative analysis to Arabidopsis (7). The EST and fatty acid metabolism sources of B. napus are available in KEGG database (8). Love et al. described a database (http://hornbill.cspp.latrobe.edu.cn) incorporating the Brassica EST, Gene Ontology (GO) annotation and information of simple sequence repeat molecular markers, of which the EST, microarray (the expression profiles of 7000 uni-genes in vegetative tissues root and leaf), and MarkerQTL information were updated (9–11), providing a useful tool for Brassica research. However, there is still no resource providing the gene expression information and the SAGE data during the reproductive development of B. napus yet. In addition, there is also no database providing bio-sources including full-length cDNA or mutants.
Based on constructing cDNA libraries using seed materials at various developmental stages, large-scale sequencing, generation of glass-based cDNA microarray and hybridization, we are able to obtain the dataset of B. napus ESTs and the relevant expression profiles during seed development, which are incorporated into Shanghai RAPESEED (RAPESEED) Database, providing a solid platform for functional genomics studies of oilseed crops. In addition, the Serial Analysis of Gene Expression (SAGE) during seed development and mutant population by ethylmethanesulfonate (EMS) mutagenesis will further facilitate the relevant studies. These will significantly enrich the relevant resources and improve the studies of the area.
SYSTEM ARCHITECTURE AND IMPLEMENTATION
RAPESEED was constructed on the platform of Sun Solaris 9 operating system and TomCat5.5 Web server. The database was implemented using a database management system MySQL5.0.20 (12). RAPESEED has been set up on a World Wide Web server allowing internet access with a web client.
SOURCES IN RAPESEED
The purpose of the RAPESEED is to provide helpful informaitons of EST, gene expression profiles and bioresources (full-length cDNA, TILLING population) to researchers and to promote the functional genomics studies and quality breeding of Brassica crops (Table 1).
Table 1.
Available resource of RAPESEED
| Resource | ||
|---|---|---|
| Unique EST | 8462; relevant expression profiles during seed development | |
| Full-length cDNA | 3526 | |
| Unique SAGE tag | 23 895 | |
| Tag-to-gene | 502 tags to B. napus | |
| 860 tags to Brassica | ||
| M3 EMS mutant individuals | ∼14 700 |
ESTs, the annotation and Gene Ontology classification
RAPESEED contains 8462 unique ESTs of B. napus, of which 6892 were functionally annotated by sequence similarity to entries within GenBank (using BLAST with a cut-off valve of E-value <10−5, 13). GO classification was performed via mapping to Uniprot to get the GO annotation. In addition, 3526 clones were full-length cDNA among the 8462 unique ESTs.
Gene expression profiles during seed development
Gene expression data for 8095 of 8462 genes during B. napus seed development are studied through cDNA microarray hybridization and available in RAPESEED, which include the expression profiles of relevant genes at 7, 9, 12, 17, 19, 21, 25, 31 day after pollination (DAP), normalized to 3 DAP (1.0).
Serial Analysis of Gene Expression (SAGE) data
A total of 23 895 unique tags from B. napus immature seeds (5 and 9 DAP) were obtained after sequencing and deposited into RAPESEED. Based on the virtual tags derived from GenBank and full-length cDNA from our cDNA library, the ‘tag-to-gene mapping database’ for B. napus and Brassica including B. oleracea and B. rapa were constructed. The whole SAGE dataset can be downloaded from RAPESEED.
EMS mutant population and seed quality measurement
The mutant population mutagenesized with ethylmethanesulfonate (EMS) was generated using B. napus (huyou-15). A population of ∼14 700 M3 individuals was generated and relevant seed qualities were measured and analyzed using the Foss NIR System. The seed quality parameters including the content of glucosinolates, protein, total fat, erucic acid, oleic acid, linoleic acid and eicosatetraenoic acid in seeds (Table 2), providing a useful source for rapeseed quality studies.
Table 2.
The summary of the seed quality measurement of EMS mutagenesized population through NIR analysis
| Glucosinolates | Protein | Lipid content | Erucic acid | Oleic acid | Linoleic acid | Eicosatetraenoic acid | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WT (Average) | 24.02 ± 2.17 | 24.74 ± 0.25 | 39.39 ± 0.42 | 7.03 ± 1.23 | 56.28 ± 1.66 | 19.62 ± 0.39 | 1.27 ± 0.77 | ||||
| WT (Maximum) | 31.37 | 25.47 | 42.43 | 9.42 | 59.91 | 21.14 | 2.97 | ||||
| WT (Minimum) | 18.99 | 21.72 | 38.26 | 3.71 | 52.86 | 18.60 | −0.49 | ||||
| Criteria | >40 | <10 | >30 | <20 | >44 | >20 | <1 | >60 | >25 | <10 | >5 |
| Numbers | 1101 | 138 | 56 | 19 | 38 | 3553 | 125 | 1254 | 432 | 117 | 365 |
Parameters of seed qualities are indicated as the content (%) in seeds. Data are presented as mean ± SD (n = 77 for wild type).
Based on the EMS mutant population, a TILLING (Targeting Induced Local Lesions IN Genomes) platform was established.
DATABASE QUERY AND USER INTERFASE
BLAST and SEARCH
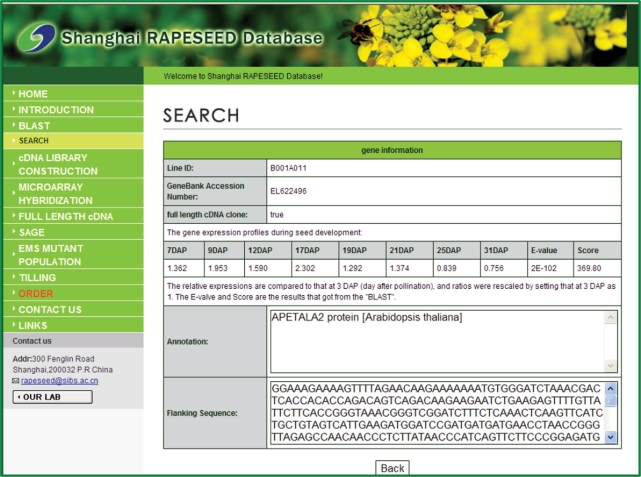
The genes of interest or EST information can be identified through the BLAST program (13). Three different BLAST programs including BLASTn, tBLASTn and tBLASTx were provided, of which the BLAST parameters can be freely chosen by users including E-value or number of alignments. The typical result page contains the collected information of the available genes in RAPESEED, including the clone ID, GenBank Accession, expression profiles during seed development, gene annotation and related E-value and score, full-length information and DNA sequences (Figure 1).
Figure 1.
Typical result page of query. The information of a gene including the clone ID, GenBank Accession, expression profiles during seed development, gene annotation and related E-value and score, full-length information and EST sequences will be shown after search.
In addition, RAPESEED provides a key word SEARCH program to facilitate the gene identification. Clone ID, GenBank Accession or key word can be selected for search. The complex SEARCH of two search items combination is also supported through and/or option.
Resource download and order from RAPESEED
All the dataset including those for whole EST sequences and corresponding annotations against GenBank, SAGE data and related tag-to-gene information, raw data of the microarray hybridization and seed qualities of EMS population (Table 2) can be downloaded. Furthermore, all listed sources of RAPESEED were available, especially the full-length cDNA clones and EMS lines with altered seed qualities, which can be ordered under the accordance of ‘Biological Material Transfer Agreement’.
AVAILABILITY
RAPESEED can be freely accessed at http://rapeseed.plantsignal.cn via the World Wide Web. We have developed a mature data management system, and all newly released information will be announced on the website. Besides ordering the interested clones or lines, users can contact us for any suggestions or questions through internet or send the comments to rapeseed@sibs.ac.cn.
FUTURE DEVELOPMENTS
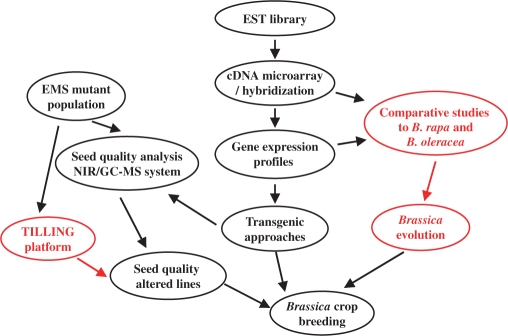
RAPESEED is a database developed for B. napus functional genomics studies, and we will do our best to continuously optimize the system and supply new records, to make it an integrated resource (Figure 2) and provide a base for Brassica research and breeding. Besides the gene identification and expression profiles, we will focus on the following in the future.
Figure 2.
Structures and development highlights of RAPESEED. The parts in red are the main directions of RAPESEED development.
TILLING population
TILLING is a useful reverse genetic technique that is a powerful, non-transgenic and unbiased targeted mutagenesis method. It can help to identify single point mutations in genes of interest (14), and is suitable for mutation site identification of the EMS lines, which is appropriate and helpful for further crop breeding. Based on the developed TILLING population of M3 EMS lines of B. napus, an amphidiploid (AACC) consisting of the genomes of B. rapa (AA) and B. oleracea (CC), we will further develop a TILLING population (>4000 M2) of diploid (B. rapa) for Brassica breeding studies. In addition, a new B. napus EMS population (M2) has already been developed this year.
We will develop pooled DNAs of ∼3000 M2 plants every year and relevant seed qualities will be measured. The screening data and seed qualities information of them will be deposited into RAPESEED annually.
Comparative gene expression profiles of B. napus to B. oleracea and B. rapa
Brassica napus (∼1200 mb) consists of that from B. rapa (700 mb) and B. oleracea (500 mb), and differences between them are obvious in many aspect such as plant morphology, seed oil content and self-productivity. This suggested that the gene expression profiles were changed dramatically during breeding processes, and investigations on these differences will surely provide the information on the regulatory network of the gene expression and the relevant cross-talk. The related information will be available soon.
ACKNOWLEDGEMENTS
The studies were supported by State Key Project of Basic Research (2006CB101603), Chinese Academy of Sciences (KSCX2-SW-328) and National Hi-Tec Program (2003AA222101). We thank Liang-Jiao Xue for his help with the database construction. Funding to pay the Open Access publication charges for this article was provided by 2006CB101603.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhang SF, Ma CZ, Zhu JC, Wang JP, Wen YC, Fu TD. Genetic analysis of oil content in Brassica napus L. using mixed model of major gene and polygene. Acta Genetica Sinica. 2006;33:171–180. doi: 10.1016/S0379-4172(06)60036-X. [DOI] [PubMed] [Google Scholar]
- 2.Zhang HZ, Shi CH, Wu JG, Ren YL, Li CT, Zhang DQ, Zhang YF. Analysis of genetic and genotype X environment interaction effects from embryo, cytoplasm and maternal plant for oleic acid content of Brassica napus L. Plant Sci. 2004;167:43–48. [Google Scholar]
- 3.Meng J, Shi S, Gan L, Li Z, Qun X. The production of yellow-seeded Brassica napus (AACC) through crossing interspecific hybrids of B. campesrtis (AA) and B. carinata (BBCC) with B. napus. Euphytica. 1998;103:329–333. [Google Scholar]
- 4.Voelker TA, Hayes TR, Cranmer AM, Turner JC, Davies HM. Genetic engineering of a quantitative trait: metabolic and genetic parameters influencing the accumulation of laurate in rapeseed. Plant J. 1996;9:229–241. [Google Scholar]
- 5.Fox SR, Hill LM, Rawsthorne S, Hills MJ. Inhibition of the glucose-6-phosphate transporter in oilseed rape (Brassica napus L.) plastids by acyl-CoA thioesters reduces fatty acid synthesis. Biochem. J. 2000;352:525–532. [PMC free article] [PubMed] [Google Scholar]
- 6.Dehesh K, Tai H, Edwards P, Byrne J, Jaworski JG. Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol. 2001;125:1103–1114. doi: 10.1104/pp.125.2.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003;31:5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Love CG, Batley J, Lim G, Robinson AJ, Savage D, Singh D, Spangenberg GC, Edwards D. New computational tools for Brassica genome research. Comput. Funct. Genomics. 2004;5:276–280. doi: 10.1002/cfg.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Love CG, Robinson AJ, Lim GAC, Hopkins CJ, Batley J, Barker G, Spangenberg GC, Edwards D. Brassica ASTRA: an integrated database for Brassica genomic research. Nucleic Acids Res. 2005;33:D656–D659. doi: 10.1093/nar/gki036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erwin TA, Jewell EG, Love CG, Lim GAC, Li X, Chapman R, Batley J, Stajich JE, Mongin E, et al. BASC: an integrated bioinformatics system for Brassica research. Nucleic Acids Res. 2007;35:D870–D873. doi: 10.1093/nar/gkl998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DuBois P. MySQL. Indianapolis, IN: New Riders; 1999. [Google Scholar]
- 13.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.McCallum CM, Comai L, Greene EA, Henikoff S. Targeted screening for induced mutations. Nat. Biotechnol. 2000;18:455–457. doi: 10.1038/74542. [DOI] [PubMed] [Google Scholar]