Abstract
High throughput methods are increasingly being used to examine the functions and interactions of gene products on a genome-scale. These include systematic large-scale proteomic studies of protein complexes and protein–protein interaction networks, functional genomic studies examining patterns of gene expression and comparative genomics studies examining patterns of conservation. Since these datasets offer different yet highly complementary perspectives on cell behavior it is expected that integration of these datasets will lead to conceptual advances in our understanding of the fundamental design and evolutionary principles that underlie the organization and function of proteins within biochemical pathways. Here we present Bacteriome.org, a resource that combines locally generated interaction and evolutionary datasets with a previously generated knowledgebase, to provide an integrated view of the Escherichia coli interactome. Tools are provided which allow the user to select and visualize functional, evolutionary and structural relationships between groups of interacting proteins and to focus on genes of interest. Currently the database contains three interaction datasets: a functional dataset consisting of 3989 interactions between 1927 proteins; a ‘core’ high quality experimental dataset of 4863 interactions between 1100 proteins and an ‘extended’ experimental dataset of 9860 interactions between 2131 proteins. Bacteriome.org is available online at http://www.bacteriome.org.
INTRODUCTION
From a historic perspective Escherichia coli has played a central role in the elucidation of the mechanisms underlying core cellular processes such as metabolism, signaling, gene expression and genome replication. A key feature of many of these processes is the tendency of their component proteins to physically associate via stable protein–protein interactions (PPI) to form larger macromolecular assemblies or complexes. These complexes are often linked together by extended networks of more transient PPI such that the cell is increasingly viewed as an assembly of interconnected functional modules—the ‘interactome’—which integrates and coordinates the cell's biochemical activities, behavior and responses to external and intrinsic signals. Systematic large-scale proteomics studies and sophisticated computational analyses are increasingly being applied to reveal the extent and complexity of these interconnections in E. coli (1–4). In addition to these interaction datasets, a large body of research has resulted in the generation of comprehensive knowledgebases providing functional and structural details of each E. coli gene product (5,6). Together with other high throughput ‘omic’ type studies measuring, for example, global patterns of gene expression (7) or the impact of evolutionary constraints (8), these complementary resources are paving the way for an exciting new era of ‘integrative biology’ where, for the first time, entire systems of interacting biomolecular components can be studied at several levels of biological abstraction. Although each dataset may be exploited for its own purposes, it is widely anticipated that close integration of these datasets will reveal a host of hitherto unknown biological relationships. For example, combining comparative genomic, pathway, structural and protein–protein interaction (PPI) data will allow the identification of not only which proteins interact, but also their overall functional organization, domain associations and evolutionary relationships.
Here we introduce a new database resource focusing on the collation of these datasets from E. coli to provide a detailed view of a model bacterial interactome (Bacteriome.org). Unlike other excellent resources which collate interaction data for a range of different organisms, for example, STRING (4), BioGRID (9) and ProLinks (2), our focus is to collate and exploit the unique properties of these complementary datasets to provide an integrated and detailed view of structural, functional and evolutionary relationships within the E. coli interactome. Two types of interaction networks are presented: an ‘experimental’ dataset that builds on a previously published high throughput protein–protein interaction screen (3); and a ‘theoretical’ dataset of predicted functional interactions constructed from the Bayesian integration of functional genomic and proteomic datasets (1). In addition to web forms allowing the interrogation and navigation of the datasets, a specialized Java applet has been created for the visualization of associated metadata such as functional categories of proteins, complex membership, protein domains and phylogenetic profiles, within the context of the interaction networks.
The database is open to browsing without restriction. Links are provided to allow users to freely download the interaction datasets.
CONSTRUCTION OF THE RESOURCE
The Bacteriome resource currently provides access to three recently derived interaction datasets for E. coli—one theoretical and two experimental (unpublished data). Detailed information on their construction and analysis is outside the scope of the current article, but is available online and will be presented in additional publications.
The first consists of a set of 3989 functional interactions predicted between 1927 proteins. These predictions were generated from the integration of a variety of experimental and computationally derived functional genomic and proteomic datasets. Sources for the experimental datasets include large- and small-scale PPI's obtained from the database of interacting proteins (DIP) (10) which includes a recently published high throughput study of E. coli PPI's (1), and co-expression data from a recent comparative study of gene expression profiles (11). Sources for the theoretical datasets include operon, gene neighborhood, gene fusion and phylogenetic profile data obtained from the Prolinks database (2); a set of interactions previously predicted from literature data (12) and a set of interactions previously predicted using the ‘interolog’ approach (13). Predictions of functional linkages between pairs of proteins were obtained using a similar naïve Bayes approach previously applied to yeast (14). In this scheme, weights are assigned to reflect the relative confidence associated with each dataset. These are derived as log likelihood scores measuring the likelihood that pairs of genes are functionally linked within a given pathway (as defined by the EcoCyc database (5)) given the evidence. Benchmarks based on: the Kyoto Encyclopaedia of Genes and Genomes (KEGG) (15); Clusters of Orthologous Genes (COG) (16); and Gene Ontology annotations (17) gave similar results. The combination of weights for an interaction identified across different datasets was then used to quantify the evidence that a given interaction is real. We used data from small-scale pull-down experiments obtained from DIP as our ‘gold standard’ set of functional linkages for determining the cutoff score for inclusion of functional linkages in the final theoretical interaction dataset. Further details including an analysis of the performance of this method are provided on the website.
The two experimental datasets represent physical interactions obtained from a high throughput screen using our previously described TAP-TAG technology (3). These include a ‘core’ dataset of 4863 interactions between 1100 proteins and an ‘extended’ dataset of 9860 interactions between 2131 proteins. For each interaction a purification enrichment (PE) score is derived which takes into account the bait_prey, prey_bait and prey_prey relationships of the interaction. Individual scores were calculated for each component based on a probabilistic discriminant function as described previously (18). The primary affinity purification scores (obtained through MS-LCMS and MALDI) and the PE scores were both used to evaluate the overall confidence of the interaction. Confidence was calculated through a logistic regression model using a weighted sum to integrate the scores (see website for further details). The two datasets were obtained using different cutoff values of their confidence scores. For the core dataset we used a confidence score cutoff of 0.7 while for the extended dataset, we used a slightly lower confidence score cutoff of 0.5.
For each interaction dataset, clusters of proteins representing functional modules (for the theoretical dataset) or protein complexes (for the experimental datasets) were predicted on the basis of their common interactions using the MCL algorithm as previously described (19). Phylogenetic profiles [representing the presence or absence of a sequence across a set of genomes (20,21)], were generated via a series of BLAST analyses (22) across 199 selected genomes (19 eukaryotes, 165 bacteria and 15 archaea).
The Bacteriome resource is implemented using postgreSQL (http://www.postgresql.org). The previously constructed E. coli knowledgebase (6) was downloaded as a set of flat files and used to build the initial resource. The additional datasets (interactions, phylogenetic profiles and predictions of protein complexes/functional modules) were imported as sets of additional tables. Users are able to browse the data via a series of php-based web pages. In addition, we have created a specialized Java applet to allow visualization and navigation of the protein networks. The applet was written using the open source Java Universal Network/Graph (JUNG) framework (http://jung.sourceforge.net/index.html).
BROWSING THE BACTERIOME
Bacteriome.org provides a number of web-based forms for querying the interaction datasets and selecting one or more proteins for either a more detailed view of the gene annotations or for viewing within the context of its interactions with other proteins: (1) Text-based searches—these include keyword searches against annotations such as gene names, protein domains, gene ontology terms and swissprot descriptions (e.g. identify all the genes which have been annotated with the term ‘kinase’); (2) Sequence similarity searches—Bacteriome.org features a BLAST page that enables users to identify E. coli homologs to their sequence of interest (e.g. identify all the genes which possess sequence similarity to protein X); (3) Phylogenetic profile searches—this allows the user to identify genes that have similar sequences in selected groups of organisms (e.g. identify all the genes which have homologs in all plants and protists); (4) Chromosomal location searches—this page allows the user to zoom in on a section of the E. coli genome and select genes on the basis of their local neighborhood (e.g. identify all genes that are within 50 kb of rpsH). (5) Browsing complexes/functional modules—finally, a Java applet is provided which allows the visualization of the predicted protein complexes/functional modules from which users may select one or more complexes for a more detailed view.
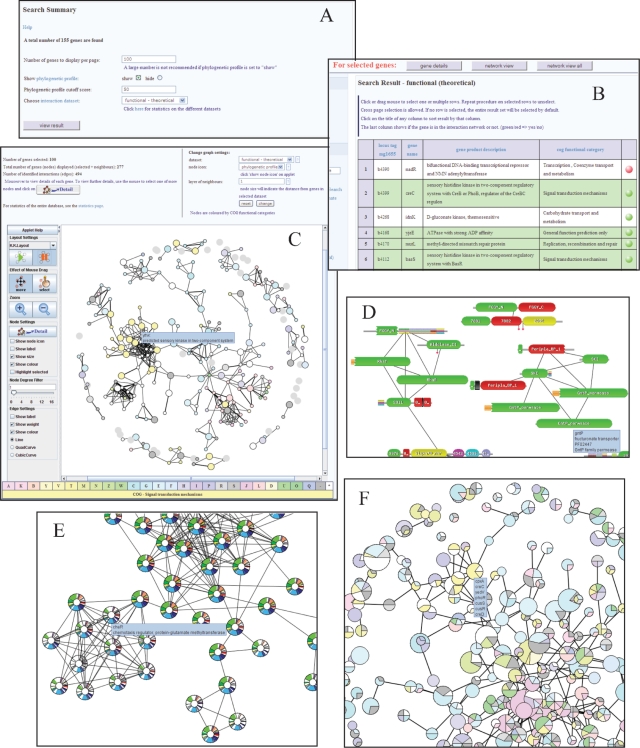
After performing a typical search (e.g. entering the term ‘kinase’ in the ‘Wild Search’ box on the left menu), the user is first presented with a summary page detailing the number of proteins matching the search (Figure 1A). In addition to formatting options, the user may select one of the three interaction datasets for subsequent network visualization. The following results page then provides the user with a list of proteins and brief descriptions (Figure 1B) from which individual, groups or even the entire dataset of proteins may be selected for either a detailed view of each protein (providing access to functional data, gene ontology terms, protein domains, sequence data and so forth) or a view of the network in which the selected protein(s) operate. The network view features a purpose built interactive Java applet in which proteins are represented by nodes in a graph (Figure 1C). The applet provides the user with a range of different layout settings and options for visualization of the network. These include the ability to navigate and zoom in on parts of the network, identifying nodes and visualizing the weights of interactions (which provide a measure of confidence). Placing the mouse over individual nodes provides details of individual proteins while a select function allows users to obtain a more detailed view of one or more nodes. The initial view of the network colors each protein (node) according to its COG functional category (16) and also displays proteins that directly interact with the initially selected proteins (the size of each node represents the distance from the initially selected proteins). However, uniquely, the applet also features the ability to change the node representations to show either the domain architecture of each protein (Figure 1D) or the phylogenetic profile of each protein (Figure 1E). Other features provided in the network view include the ability to alter the layer of neighbors presented in the network (e.g. nearest neighbors to the selected proteins, next nearest neighbors to the selected proteins) and the ability to choose which interaction dataset to visualize.
Figure 1.
Typical screenshots from Bacteriome.org. (A) Summary page of a typical search. Here we have identified 155 genes associated with the word ‘kinase’ that was entered in the wild search box on the home page. The user may select one of the three datasets to view interactions associated with these 155 genes. (B) Search results pages. These pages provide summary information on each gene identified by a search. One or more genes may be selected for either a more detailed view of each gene or for viewing within the context of an interaction network. An additional button is provided to view the network of all identified genes. (C) Network view. The embedded java applet provides an interactive view of the interactions associated with 100 selected genes (large nodes). In addition to switching between different settings such as the interaction dataset and layers of neighbors to view, the Java applet features a graphical user interface to manipulate the network view. For example, the user could zoom into a section of the network, select and move groups or individual proteins and choose to view the nodes in terms of their PFAM domain architecture. (D) Alternatively, the user could also view the nodes in terms of their phylogenetic profiles. (E) The presented example shows the profiles for a group of chemotaxis related proteins that appear to form a functional module (left). Note how many of the proteins in this module appear to have homologs in a few restricted taxonomic groups including the various proteobacteria groups (different shades of blue), spirochaetes (purple), firmicutes (green), cyanobacteria (yellow) and archaea (red) suggesting a degree of evolutionary modularity. (F) In addition to visualizing interactions between individual proteins, the Java applet has also been adapted to provide a view of predicted protein complexes/functional modules. This view shows a section of the interactions between the functional modules predicted for the functional interaction network. Each pie chart shows the proportion of proteins associated with each COG functional category. The size of the pie indicates the number of proteins associated with each complex/module. Placing the mouse over the pie provides details of constituent proteins which can be selected for a more detailed view.
Browsing the experimental protein complexes or the theoretical functional modules associated with the networks takes the user directly to a network view of the complexes/modules in which each node (representing a complex/module) is visualized as a pie chart showing the proportion of proteins in the complex/module associated with particular COG functional categories (Figure 1F). Here, the size of each node indicates the number of protein constituents, details of which may be obtained through placing the mouse over the node in question. Again, users may select individual or groups of nodes for a more detailed report of the associated proteins (including the ability to visualize their local network).
FUTURE DIRECTIONS
We are continuing to generate new physical interaction data for E. coli and in the near future we hope to have completed interaction mapping for at least three quarters of E. coli proteins. These datasets together with updated predictions of protein complexes will be integrated in the Bacteriome resource as they are generated. We are also planning to host additional experimental and theoretical bacterial interaction datasets such as the yeast two-hybrid datasets for Helicobacter pylori (23) and Campylobacter jejuni (24). The inclusion of these datasets will necessitate the creation of corresponding knowledgebases providing detailed functional and structural annotations. These will be developed using the existing resource for E. coli (6) as a template. Aside from the interaction datasets, we are also seeking to extend the types of metadata that may be incorporated into the resource. These might include expression datasets (7) in which the expression pattern of a protein under a set of conditions could be visualized within a network setting using pie charts in an analogous fashion to that implemented by the GenePro plugin for Cytoscape (25,26).
ACKNOWLEDGEMENTS
This work was funded by the Canadian Institute of Health Research (CIHR). J.P. is supported by a New Investigators award from CIHR. Funding to pay the Open Access publication charges for the article was provided by CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Arifuzzaman M., Maeda M., Itoh A., Nishikata K., Takita C., Saito R., Ara T., Nakahigashi K., Huang H.C., et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowers P.M., Pellegrini M., Thompson M.J., Fierro J., Yeates T.O., Eisenberg D. Prolinks: a database of protein functional linkages derived from coevolution. Genome Biol. 2004;5:R35. doi: 10.1186/gb-2004-5-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butland G., Peregrin-Alvarez J.M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 4.von Mering C., Jensen L.J., Kuhn M., Chaffron S., Doerks T., Kruger B., Snel B., Bork P. STRING 7–recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keseler I.M., Collado-Vides J., Gama-Castro S., Ingraham J., Paley S., Paulsen I.T., Peralta-Gil M., Karp P.D. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riley M., Abe T., Arnaud M.B., Berlyn M.K., Blattner F.R., Chaudhuri R.R., Glasner J.D., Horiuchi T., Keseler I.M., et al. Escherichia coli K-12: a cooperatively developed annotation snapshot–2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faith J.J., Hayete B., Thaden J.T., Mogno I., Wierzbowski J., Cottarel G., Kasif S., Collins J.J., Gardner T.S. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen L., Bollback J.P., Dimmic M., Hubisz M., Nielsen R. Genes under positive selection in Escherichia coli. Genome Res. 2007;17:1336–1343. doi: 10.1101/gr.6254707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergmann S., Ihmels J., Barkai N. Similarities and differences in genome-wide expression data of six organisms. PLoS Biol. 2004;2:E9. doi: 10.1371/journal.pbio.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann R., Valencia A. Implementing the iHOP concept for navigation of biomedical literature. Bioinformatics. 2005;21(Suppl. 2):ii252–ii258. doi: 10.1093/bioinformatics/bti1142. [DOI] [PubMed] [Google Scholar]
- 13.Yu H., Luscombe N.M., Lu H.X., Zhu X., Xia Y., Han J.D., Bertin N., Chung S., Vidal M., et al. Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee I., Date S.V., Adai A.T., Marcotte E.M. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- 15.Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins S.R., Kemmeren P., Zhao X.C., Greenblatt J.F., Spencer F., Holstege F.C., Weissman J.S., Krogan N.J. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 20.Pellegrini M., Marcotte E.M., Thompson M.J., Eisenberg D., Yeates T.O. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcotte E.M., Pellegrini M., Ng H.L., Rice D.W., Yeates T.O., Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Rain J.C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 24.Parrish J.R., Yu J., Liu G., Hines J.A., Chan J.E., Mangiola B.A., Zhang H., Pacifico S., Fotouhi F., et al. A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 2007;8:R130. doi: 10.1186/gb-2007-8-7-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vlasblom J., Wu S., Pu S., Superina M., Liu G., Orsi C., Wodak S.J. GenePro: a Cytoscape plug-in for advanced visualization and analysis of interaction networks. Bioinformatics. 2006;22:2178–2179. doi: 10.1093/bioinformatics/btl356. [DOI] [PubMed] [Google Scholar]
- 26.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]