Abstract
The knowledge about interactions between proteins and small molecules is essential for the understanding of molecular and cellular functions. However, information on such interactions is widely dispersed across numerous databases and the literature. To facilitate access to this data, STITCH (‘search tool for interactions of chemicals’) integrates information about interactions from metabolic pathways, crystal structures, binding experiments and drug–target relationships. Inferred information from phenotypic effects, text mining and chemical structure similarity is used to predict relations between chemicals. STITCH further allows exploring the network of chemical relations, also in the context of associated binding proteins. Each proposed interaction can be traced back to the original data sources. Our database contains interaction information for over 68 000 different chemicals, including 2200 drugs, and connects them to 1.5 million genes across 373 genomes and their interactions contained in the STRING database. STITCH is available at http://stitch.embl.de/
INTRODUCTION
In pharmacology and biochemistry the interplay of chemicals and proteins has been studied over many years, but much of the existing data on chemicals is either hidden in a vast amount of dispersed literature or is locked away in commercial databases such as the Chemical Abstracts Service Registry. Recently, however, several projects have begun to provide easy public access to chemical information. Resources such as PubChem (1), ChEBI (2) and ChemDB (3) provide an ever-growing inventory of the chemical space that can be used as the basis for the integration of knowledge about chemicals themselves, their biological interactions and their phenotypic effects. Thus, many problems in Chemical Biology are now becoming approachable by the academic research community.
Valuable information about the biological activity of chemicals is provided by large-scale experiments. Phenotypic effects of chemicals were first made available on a large scale by the US National Cancer Institute (NCI), which conducts anti-cancer drug screens on 60 human tumour cell lines (NCI60) (4). The patterns of growth inhibition in the different cell lines by small molecules can not only be used to judge the efficacy of individual compounds, but also to relate compounds by their mechanism of action (5,6). Other unexpected relationships between compounds can be found using the PubChem BioAssay resource, where NCI60 data and many other assays are aggregated. As of July 2007, it contains 587 highly diverse assays, ranging from studies of single molecules to high-throughput screens with over 100 000 tested substances. Recently, the Connectivity Map project (7) set out to catalogue the perturbations in gene expression upon chemical treatment. As the Connectivity Map increases in coverage of small molecules, it will develop into a very useful resource for both the similarities of chemicals and the relations of chemicals with proteins.
Information about interactions between proteins and small molecules is essential for understanding metabolism, signalling and drug treatment. Part of this information is stored in different types of databases. There are some databases that focus on the biological actions of drugs, for example DrugBank (8), TTD (9), SuperTarget and MATADOR (10). Another database, the PDSP Ki Database (11) provides protein binding constants (Ki) for compounds, combining data from the literature and internal screens. Finally, there are many pathway databases most prominently KEGG (12), MetaCyc (13) and Reactome (14). These diverse knowledge sources can be integrated to provide links between chemical space and the protein universe through genome databases such as Ensembl (15) and RefSeq (16).
In order to get a complete picture of the biochemistry of metabolites, drugs and other compounds, the current challenge is to integrate the various sources of chemical knowledge into a single resource and to link it with the knowledge about proteins. For example, to be able to link phenotypic observations from cell line screens to molecular events, their interactions with proteins in the context of cellular networks are essential.
Here, we present search tool for interactions of chemical (STITCH), a search tool and resource for the interactions of chemicals and proteins. A consolidated set of chemicals is derived from PubChem. Relations between chemicals are derived from similar activity profiles in the NCI60 cell lines, from pharmacological actions assigned to chemicals in the Medical Subject Headings (MeSH) and from the literature. Chemical–chemical and chemical–protein associations are integrated from pathway and experimental databases, as well as from the literature. Lastly, as many associations as possible are annotated with interaction types.
CONSOLIDATED SET OF CHEMICALS
Chemicals are the basis of STITCH and are currently imported from PubChem. All stereoisomers and charge forms of a compound are merged into one record via the canonical SMILES string. While this might be an over-simplification, it is necessary and valid for three reasons: Stereoisomers often share names, for example the name ‘valine’ is assigned to l-valine, d-valine and a third compound without stereochemistry and therefore, it is not possible to automatically assign the synonym. Second, external databases may link to the compound with or without stereochemistry. Lastly, enantiomers with different biological activity may interconvert in vivo. This is the case for the drug thalidomide, where one enantiomer can be used to treat morning sickness, but the other enantiomer causes birth defects (17).
Drugs are often marketed as different salts and mixtures of the same active substance, which are represented as distinct entries in the chemical databases. As the different formulations will have the same biological effect, they are joined into one entry in STITCH. In order to do this, a list of 30 additive compounds that can be discarded was created by manual inspection of mixtures. This list includes water, ions such as calcium and organic acids such as methanesulfonic acid (which forms mesylates). When these compounds are present together with an organic compound of at least 100 g/mol, the additive compounds are discarded and the database entry of the mixture is joined to the entry of the base compound. For example, imatinib mesylate is reduced to its main form imatinib.
The PubChem database is an aggregation of many databases. Often, different databases contradict each other in the assignment of synonyms to compounds. For this reason, some of the source databases have been identified as either trusted or dubious sources. This information is used to arbitrate conflicts in the assignment of synonyms.
ASSOCIATIONS BETWEEN CHEMICALS
Building upon the set of chemicals, associations of chemicals and proteins can be imported from various sources. Taken together, these associations form a network that can be used to explore the context of chemicals and proteins. The associations derived here are combined with protein–protein interactions stored in the STRING database (18) to form one large network.
Four types of edges link chemicals in the chemical–chemical network: reactions from pathway databases, literature associations, similar structures and similar activities. Pathway databases contain records about chemical reactions that are used to derive associations. The open-source Chemistry Development Kit (19) was used to calculate chemical fingerprints and the commonly used Tanimoto 2D chemical similarity scores (20,21). Literature associations were derived in the same manner as chemical–protein associations (see subsequently).
To predict whether two chemicals have similar molecular activities, data from MeSH pharmacological actions and activities in the NCI60 screens were used. Compounds from MeSH were mapped by their name to the database of chemicals. To assess the relevance of the association by a shared pharmacological action, the number of chemicals annotated with each pharmacological action is determined and benchmarked against the probability of sharing a drug target in the MATADOR database.
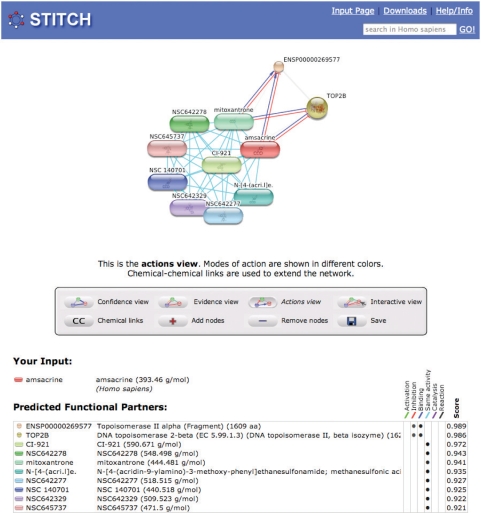
An activity pattern was calculated for each compound based on the NCI60 screens, by converting its log(GI50) values (the concentrations required to inhibit growth by 50%) to Z-scores in each cell line (6). Compounds with uninformative activity patterns were excluded using a threshold for the standard deviation of the Z-scores. Then, the Pearson correlation of the activity patterns was calculated for all compound pairs and benchmarked against known mechanisms of action (6,22). This allows the user to link compounds with unknown mechanism of action to well-studied compounds (Figure 1).
Figure 1.
Interactions of the topoisomerase II inhibitor amsacrine. Chemicals are represented as pill-shaped nodes, while proteins are shown as spheres. Nodes that are associated to each other are linked by an edge: like mitoxantrone, amsacrine is known to bind (blue lines) and inhibit (red arrows) type II topoisomerases. Other compounds have similar activities as amsacrine or mitoxantrone in the NCI60 anti-cancer drug screens and are thus predicted to have the same mechanism of action (cyan lines).
SOURCES OF CHEMICAL–PROTEIN INTERACTIONS
In order to link the derived chemical–chemical associations to the protein world, a variety of databases of chemical–protein interactions are imported. Experimental evidence of direct chemical–protein binding is derived from the PDSP Ki Database (11) and the protein data bank (PDB) (23). Additional interactions between metabolites and proteins are extracted from the manually annotated pathway databases such as KEGG (12), Reactome (14) and the NCI-Nature Pathway Interaction Database (http://pid.nci.nih.gov), and drug–target relations are imported from DrugBank (8) and MATADOR (10).
Text mining of MEDLINE and OMIM yields additional evidence, based both on a simple co-occurrence scheme and a more complex natural language processing (NLP) approach (24,25). In order to increase the coverage of the text-mining approach, groups of proteins that are described in MeSH terms are also used as entities during text mining. This allows us to capture interactions such as the binding of memantine to multimeric NMDA (N-methyl d-aspartate) receptors. The MeSH terms are mapped to proteins by the identifiers and synonyms provided by MeSH. If this automatic mapping is not possible, common proteins were manually assigned to candidate MeSH terms that were determined by using the annotations of abstracts with MeSH terms in MEDLINE. To reflect the decreased confidence associated with an interaction of a compound with a group of proteins rather than a single protein, the interaction score was scaled down as a function of the sequence diversity within the group.
For each individual evidence type, likelihood or relevance scores have been developed. The individual scores for a given chemical–protein or chemical–chemical interaction are then combined into one overall score (26). Chemical–protein interactions are transferred between species based on the sequence similarity of the proteins (26).
In order to be as comprehensive as possible, STITCH derives data from a variety of sources. While the sources have different focuses, it is well possible that certain areas of knowledge are not covered. In the future, more data sources will be added to further increase the coverage of STITCH.
ANNOTATION OF SMALL MOLECULE ACTIONS
Many data sources contain information about the biological or biochemical action associated with a certain interaction. This information can be stated explicitly, like in databases using the BioPAX ontology (27), or implicitly, like in crystal structures. As one of the display modes, STITCH allows the user to view a network of interactions augmented by the types of actions (Figure 2). Possible actions are: activation, inhibition, direct binding, catalysis, (bio)chemical reaction and similar activity. To avoid overloading the visual network representation with a great number of edge types, this representation is intentionally not as detailed as provided by the BioPAX ontology or recently suggested by Lu et al. (28).
Figure 2.
Network around acetylcholine and acetylcholinesterase (ACHE). (a) In confidence view, thicker lines represent stronger associations. (b) Lines and, for directed edges, arrows of different colours stand for different edge types in the actions view: binding (blue), activation (green), inhibition (red), catalysis (magenta), same activity (cyan) and reaction (black). The network shows the hydrolysis from acetylcholine to choline that is catalysed by ACHE. Several drugs, for example, the nootropic drug donepezil, inhibit ACHE.
Taken together, STITCH links molecular, cellular and phenotypic data related to small molecules and allows easy navigation in and visualization of networks of large collections of associations between chemicals as well as interactions between chemicals and proteins. It thus represents a useful resource for both in-depth and large-scale projects in Chemical Biology.
DATABASE ACCESS AND NETWORK VIEWS
Users can query the STITCH database (http://stitch.embl.de) in several ways. A full-text search is available for identifiers and common names of chemicals and proteins. Chemical structures may be entered as SMILES strings to search for similar chemicals that are stored in the database. Finally, protein sequences can be submitted to find similar proteins in the database.
When searching STITCH with a chemical as entry point, the user is presented with a network of related proteins that places the chemical into a biological context. The network can be extended to also show related chemicals, which is useful for highlighting, for example, compounds with similar pharmacological activity or metabolized forms. Querying STITCH for a protein will provide the user with a network that places the protein into its chemical and biological context. The network viewer displays chemical and protein structures and provides the user with easy access to information from resources such PubChem (1), PDB (23) and SMART (29).
To aid the user in exploring and interpreting the networks, we provide four different views. By default, the confidence view is shown (Figures 1 and 2a). There the thickness of the lines represents the confidence score of the association. In the evidence view, separate lines with different colours are used to show the type of evidence that support each interaction, for example experimental evidence or text mining. Where possible, the actions view uses different colours to visualize the types of interaction between chemicals and proteins, for example activation, inhibition or metabolization (Figure 2b). Finally, an interactive view allows the user to modify the network layout.
For large-scale analyses, the interaction network of chemicals and proteins can be downloaded from the STITCH website. We also make available synonyms lists for chemicals and proteins as well as a set of database cross-references to other chemical databases. These files are available under a Creative Commons Attribution 3.0 License.
ACKNOWLEDGEMENTS
The authors thank members of the Bork and von Mering groups for comments on the STITCH web interface. This work was funded by the BMBF QuantPro grant (0313831D). Funding to pay the Open Access publication charges for this article was provided by the European Molecular Biology Laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooksbank C, Cameron G, Thornton J. The European Bioinformatics Institute's data resources: towards systems biology. Nucleic Acids Res. 2005;33:D46–D53. doi: 10.1093/nar/gki026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen JH, Linstead E, Swamidass SJ, Wang D, Baldi P. Bioinformatics. 2007. ChemDB update—full-text search and virtual chemical space. doi: 10.1093/bioinformatics/btm1341. [DOI] [PubMed] [Google Scholar]
- 4.Weinstein JN, Myers TG, O’Connor PM, Friend SH, Fornace AJ, Kohn KW, Fojo T, Bates SE, Rubinstein LV, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 5.Huang R, Wallqvist A, Thanki N, Covell DG. Linking pathway gene expressions to the growth inhibition response from the National Cancer Institute's anticancer screen and drug mechanism of action. Pharmacogenomics J. 2005;5:399. doi: 10.1038/sj.tpj.6500331. [DOI] [PubMed] [Google Scholar]
- 6.Rabow AA, Shoemaker RH, Sausville EA, Covell DG. Mining the National Cancer Institute's tumor-screening database: identification of compounds with similar cellular activities. J. Med. Chem. 2002;45:840. doi: 10.1021/jm010385b. [DOI] [PubMed] [Google Scholar]
- 7.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 8.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Ji ZL, Chen YZ. TTD: therapeutic target database. Nucleic Acids Res. 2002;30:415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Günther S, Kuhn M, Dunkel M, Campillos M, Senger C, Petsalaki E, Ahmed J, Urdiales EG, Gewiess A, et al. SuperTarget and matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkm862. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roth B, Lopez E, Patel S, Kroeze W. The multiplicity of serotonin receptors: uselessly diverse molecules or an embarrassment of riches? Neuroscientist. 2000;6:262. [Google Scholar]
- 12.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caspi R, Foerster H, Fulcher CA, Hopkinson R, Ingraham J, Kaipa P, Krummenacker M, Paley S, Pick J, et al. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2006;34:D511–D516. doi: 10.1093/nar/gkj128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, et al. Reactome: a knowledge base of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eriksson T, Björkman S, Roth B, Fyge A, Höglund P. Stereospecific determination, chiral inversion in vitro and pharmacokinetics in humans of the enantiomers of thalidomide. Chirality. 1995;7:52. doi: 10.1002/chir.530070109. [DOI] [PubMed] [Google Scholar]
- 18.von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Krüger B, Snel B, Bork P. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinbeck C, Hoppe C, Kuhn S, Floris M, Guha R, Willighagen EL. Recent developments of the chemistry development kit (CDK)—an open-source java library for chemo- and bioinformatics. Curr. Pharm. Des. 2006;12:2120. doi: 10.2174/138161206777585274. [DOI] [PubMed] [Google Scholar]
- 20.Martin YC, Kofron JL, Traphagen LM. Do structurally similar molecules have similar biological activity? J. Med. Chem. 2002;45:4358. doi: 10.1021/jm020155c. [DOI] [PubMed] [Google Scholar]
- 21.Willett P, Barnard JM, Downs GM. Chemical similarity searching. J. Chem. Inf. Comput. Sci. 1998;38:983–996. [Google Scholar]
- 22.Weinstein JN, Kohn KW, Grever MR, Viswanadhan VN, Rubinstein LV, Monks AP, Scudiero DA, Welch L, Koutsoukos AD, et al. Neural computing in cancer drug development: predicting mechanism of action. Science. 1992;258:447–451. doi: 10.1126/science.1411538. [DOI] [PubMed] [Google Scholar]
- 23.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen L, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat. Rev. Genet. 2006;7:129. doi: 10.1038/nrg1768. [DOI] [PubMed] [Google Scholar]
- 25.Saric J, Jensen LJ, Ouzounova R, Rojas I, Bork P. Extraction of regulatory gene/protein networks from Medline. Bioinformatics. 2006;22:645–650. doi: 10.1093/bioinformatics/bti597. [DOI] [PubMed] [Google Scholar]
- 26.von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stromback L, Lambrix P. Representations of molecular pathways: an evaluation of SBML, PSI MI and BioPAX. Bioinformatics. 2005;21:4407. doi: 10.1093/bioinformatics/bti718. [DOI] [PubMed] [Google Scholar]
- 28.Lu LJ, Sboner A, Huang YJ, Lu HX, Gianoulis TA, Yip KY, Kim PM, Montelione GT, Gerstein MB. Comparing classical pathways and modern networks: towards the development of an edge ontology. Trends Biochem. Sci. 2007;32:310–321. doi: 10.1016/j.tibs.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]