Abstract
MicroRNAs (miRNAs) constitute an important class of regulators that are involved in various cellular and disease processes. However, the functional significance of each miRNA is mostly unknown due to the difficulty in identifying target genes and the lack of genome-wide expression data combining miRNAs, mRNAs and proteins. We introduce a novel database, miRGator, that integrates the target prediction, functional analysis, gene expression data and genome annotation. MiRNA function is inferred from the list of target genes predicted by miRanda, PicTar and TargetScanS programs. Statistical enrichment test of target genes in each term is performed for gene ontology, pathway and disease annotations. Associated terms may provide valuable insights for the function of each miRNA. For the expression analysis, miRGator integrates public expression data of miRNA with those of mRNA and protein. Expression correlation between miRNA and target mRNA/proteins is evaluated and their expression patterns can be readily compared. Our web implementation supports diverse query types including miRNA name, gene symbol, gene ontology, pathway and disease terms. Interfaces for exploring common targets or regulatory miRNAs and for profiling compendium expression data have been developed as well. Currently, miRGator, available at: http://genome.ewha.ac.kr/miRGator/, supports the human and mouse genomes.
INTRODUCTION
MicroRNAs (miRNAs), a family of small noncoding RNAs of ∼22 nt in length, constitute an important class of regulators that are involved in diverse cellular processes such as developmental control, apoptosis, cell differentiation and proliferation (1). They are also implicated in various disease processes thus emerging as potential targets of therapeutic intervention (2,3).
Significant efforts have been made to identify miRNAs and their target mRNAs during last several years. Sanger Institute's miRBase serves as the central depository of miRNAs that are experimentally validated (4). The current release, version 10.0, contains over 5000 miRNAs from various organisms including 533 human and 442 mouse miRNAs. However, the function of each miRNA is mostly unknown except a few miRNAs so far, and diverse experimental and computational approaches are being applied to elucidate their functional significance (5,6).
MiRNAs are involved in the regulation of protein expression primarily by binding to one or more target sites on an mRNA transcript and inhibiting translation. Thus, identification of target mRNAs is of utmost importance aspect in understanding miRNA function. Computational prediction of target genes in animal has proven challenging mainly due to imperfect base pairing and the limited length of binding sites (7). PicTar (8) and TargetScanS (9) are two prominent programs that utilize cross-species conservation and the near-perfect complementarity between the 5' seed region of miRNA and the binding sites of target mRNA for the prediction of target mRNAs. Their genome-wide analysis results are available in the UCSC genome browser database (10). Also of utility is Tarbase which is a manually curated collection of experimentally tested miRNA targets in eight organisms (11).
Recent databases on miRNAs tend to combine the compilation of miRNA with target prediction modules. miRBase has added the target prediction feature as well using the miRanda algorithm (12). Argonaute provides compiled information on miRNAs of human, mouse and rat (13). miRNAMap offers an enhanced interface for known and predicted miRNAs (14). Argonaute and miRNAMap provide the expression profiles of known miRNAs although the coverage to date is rather limited. miRGen is a new addition to this list with many convenient features (15). It supports more organisms, diverse types of gene id, and most of the leading target prediction programs including miRanda, PicTar, TargetScanS, DIANA-microT (16). A unique interface allows users to find the clusters of miRNAs at any interval of chromosomes.
Still, the current state is that most databases available in public so far are simple collection of miRNA-related information such as miRNA itself and target binding. No systematic approach has been made for functional interpretation of miRNA targets. Even though several databases include expression information of miRNAs, the coverage is quite limited, failing to integrate most of the high-throughput experimental results (11,13).
Here, we introduce an integrated database and web interface for functional annotation of miRNAs that encompasses expression, function, pathway, disease terms as well as miRNA targeting. Three prediction programs (miRanda, PicTar and TargetScanS) are used for target prediction and their result may be combined in a Boolean logic.
Mechanistic understanding of miRNA functions has relied on the properties of a single key target gene in a regulatory or signaling pathway so far. However, since an animal miRNA is expected to target several hundred genes on average (9), it is possible that a single miRNA targets several genes of related functions. Even though it remains to be determined whether or not simultaneous targeting of related genes is the norm and provides a coordinated control mechanism as often seen in the transcriptional regulation, functional relationship between target genes is certainly valuable information in exploring functional significance of each miRNA. In this context, miRGator provides a utility for statistical enrichment test of miRNA targets in a number of annotation categories such as the gene ontology (GO) function (17), GenMAPP and KEGG pathways (18,19), and various diseases.
Expression profile of miRNAs is an important part of functional annotation and we imported the miRNA-related expression data from the gene expression omnibus (GEO) database (20). Several reports have described correlated expression of miRNAs and their target genes (21,22). Expression pattern of each miRNA can be readily compared with that of target mRNAs and proteins. Importantly, expression correlation between two types of data is also calculated as an estimate for the effect of miRNA binding. Reciprocal expression pattern between miRNA and mRNA/protein can be an indirect evidence of miRNA targeting. Web implementation supports diverse workflows that include search by miRNA(s) or target gene(s), search by functional categories and expression profiling of miRNAs in GEO. miRGator thus serves as a comprehensive resource for exploring functional aspects of miRNAs.
IMPLEMENTATION AND DESIGN
Overview and design principle
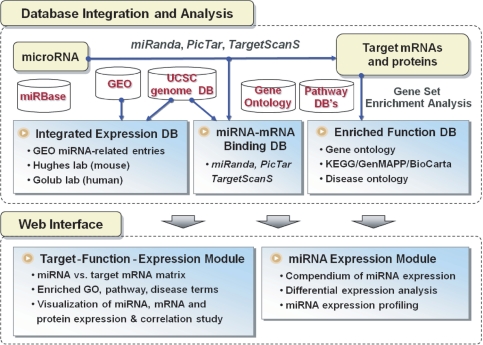
Figure 1 shows the overview of database integration, analysis flow and web implementation of miRGator. Functional analysis begins with target prediction. We used the miRanda, PicTar and TargetScanS programs whose genome-wide predictions can be downloaded from the miRBase website and the UCSC genome browser tracks. The lists of target mRNAs are tested for statistical over-representation in any functional nodes using hypergeometric distribution of Fisher's exact test. Implemented functional categories include the GO, KEGG/GenMAPP/BioCarta pathways (18,19) and disease ontology of Ingenuity Pathway Analysis.
Figure 1.
Schematic overview of miRGator system.
Since gene expression is an important part of functional annotation, we integrated various miRNA-related expression data from the GEO database and built a compendium of miRNA expression data in a similar fashion to the Oncomine cancer profiling database (23). Each data set was analyzed for differentially regulated miRNAs after quantile normalization. Simple interface was developed to visualize the expression profile of miRNAs and to address the issue of differential regulation in various situations.
Prediction of target transcripts often yields false targets even with the state-of-the-art algorithms due to imperfect base pairing and the short length of binding sites. Examining expression correlation of miRNA and the predicted target mRNAs/proteins may provide clues of genuine targeting or indirect regulation. We collected genome-wide expression data of miRNA, mRNA and protein for matching tissues/samples and evaluated the expression correlation coefficients for all miRNA–target pairs.
MicroRNA binding to target mRNA, a bipartite relationship in graph theory, is represented as a ‘miRNA–target mRNA table’ whose element is the number of binding sites. Advantages of table representation become obvious when multiple miRNAs and target genes are simultaneously examined. For example, co-regulation by multiple miRNAs can be easily explored by examining common targets. Candidate regulatory miRNAs can be obtained by providing list of genes from pathway databases or from microarray clustering results.
The concepts of miRNA targeting, functional enrichment analyses and expression correlation are closely related subjects. We built an integrated module with diverse biological questions into consideration as described in later section. All results are pre-calculated and stored in the database to speed-up the response.
Data sources and methods
Many databases of diverse characteristics are closely integrated in miRGator as listed in Figure 1. Summary of the target prediction programs is given in Table 1. Note that the degree of cross-species conservation is different between programs. The UCSC genome maps of the NCBI Build 35 (hg17) and the NCBI Build 35 (mm7) were used for the human and mouse genomes, respectively. Genome-wide prediction results from the PicTar and TargetScanS programs were obtained from the UCSC genome browser database. Target genes from miRanda 4.0 were obtained from the miRBase website where the most up-to-date information were available. Downloaded targets on the current genomes (hg18 and mm8) were lifted back to the previous genomes (hg17 and mm7), which substantially increases the miRNA coverage.
Table 1.
Statistics for various target prediction methods
| Human (hg17) | Mouse (mm7) | ||||||
|---|---|---|---|---|---|---|---|
| miRanda | PicTar-4way | PicTar-5way | TargetScanS | miRanda | PicTar-dog | PicTar-chicken | |
| Number of miRNAs | 470 | 179 | 131 | 139 | 375 | 269 | 249 |
| Number of target genes | 15 274 | 9152 | 3455 | 7709 | 14 768 | 6550 | 1492 |
| Number of binding sites | 284 714 | 154 894 | 28 870 | 22 837 | 241 791 | 106 022 | 8354 |
| Average number of target genes per miRNA | 32.5 | 51.1 | 26.4 | 55.5 | 39.4 | 24.3 | 6.0 |
| Average number of binding sites per miRNA | 606 | 865 | 220 | 164 | 645 | 394 | 34 |
| Average number of binding sites per gene | 18.6 | 16.9 | 8.3 | 3.0 | 16.4 | 16.2 | 5.7 |
Note: Cross-species conservation for each prediction method: miRanda (version 4.0): conserved in at least two species PicTar-4way: conserved in 4 species (human, mouse, rat, dog) PicTar-5way: conserved in 5 species (human, mouse, rat, dog, chicken) TargetScanS: conserved in 5 species (human, mouse, rat, dog, chicken) PicTar-dog: conserved in 7 species (mouse, rat, rabbit, human, chimp, macaque, dog) PicTar-chicken: conserved in 13 species (7 species + cow, armadillo, elephant, tenrec, opossum, chicken).
Genome annotation data, mapping genes to nodes of functional classification system, were collected from various resources. Gene-to-GO mapping was achieved by combining the UCSC kgXref table (known gene to UniProt ID) and GOA association table (UniProt ID to GO nodes) from the GO web site (10,24). Genes in the KEGG/GenMAPP/BioCarta pathways were obtained from ArrayXPath database (25). IPA's gene to disease mapping from Ingenuity Systems was used to test disease enrichment of miRNA targets. IPA's disease classification system consists of more than 7000 terms organized in three hierarchical levels of depth.
Expression data for correlation study are based on two major sources. Hughes and coworkers (26) generated a series of genome-wide expression data for mouse genome using homogeneous samples. MiRNA microarray data are available for 78 miRNAs in 17 tissues. Their mRNA expression data cover 55 tissues (27) and the proteomic data include 4768 proteins in six organs (28). As for the human genome, Golub and coworkers published expression profile of mRNA and 217 miRNAs in 334 samples (3,29). No global proteomic data in multiple tissues are available for human to the best of our knowledge, and we simply compared the expression profiles of miRNA and mRNA. Thus, the expression correlation analysis for mouse covers miRNA, mRNA, and proteins, whereas only the expression correlation between miRNA and mRNA is available for human.
We also built a compendium of miRNA expression data. Twelve miRNA-related datasets (566 samples) were downloaded from the GEO database. Proper normalization process would ideally take the unique features/characteristics of dataset into consideration. Analyzing compendium datasets, however, requires a uniform normalization procedure for convenience in implementation. We used the quantile normalization that performed best in the Affymetrix arrays (30) since most miRNA microarrays are single channeled (3). Each dataset was manually examined to set up 106 two-class comparison studies to find differentially expressed miRNAs in various situations. Seven studies compared cancer and normal tissues for bladder, breast, colon, eye, kidney, lung and uterus. Most other studies were designed to find the tissue-specific or cell-type-specific miRNAs.
USER INTERFACE
Target–function–expression module
This is the main interface of miRGator for examining target genes, inferred functions and the correlated expression through target prediction. Available target prediction methods and statistics are summarized in Table 1. Default choice is miRanda 4.0 from the miRBase since it covers the most recent compendium of miRNAs. Other methods are rather outdated with lower coverage (genome-wide calculation performed almost 2 years ago) but their target sites are conserved in more species, which may be of help in filtering out false positives. Average number of target genes and binding sites in Table 1 would be helpful in estimating reliability of prediction methods.
Each method produces a different list of target genes and it is often desirable to compare the contents. We support the Boolean combination of target gene lists from different methods. Since the miRNA coverage and the extent of conservation are different among prediction methods, we constructed a target summary table that showed the number of target genes for all miRNAs according to the prediction methods. This table can be used to pre-examine the number of target genes before actual query. Clicking on each number in the table opens up the list of target genes for the prediction method of choice.
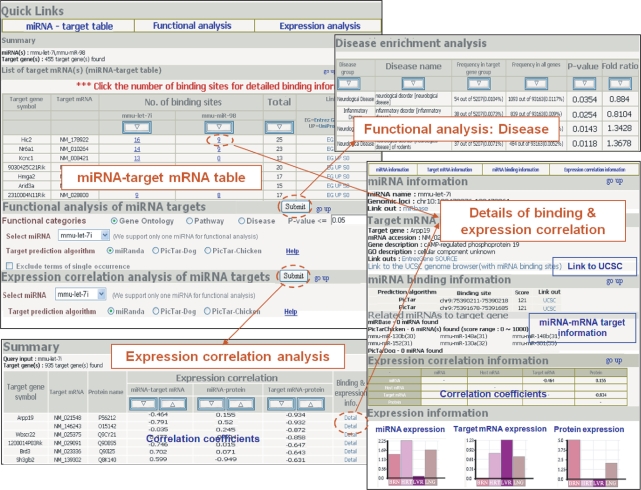
The main search can be initiated either with miRNA(s) or with target gene(s). Figure 2 is the collection of screenshots from the target–function–expression module. It consists of three major parts of miRNA–target mRNA table, functional enrichment analysis of target genes, and expression correlation analysis of target genes. Search result is always displayed in the miRNA–target table format where the number of binding sites is indicated. Clicking on each number in the table leads to the detailed information on target binding and the expression correlation pattern for corresponding miRNA–mRNA pair. We support sorting target mRNAs according to the number of binding sites, which would allow users to concentrate on mRNAs with multiple binding sites preferentially. Another advantage is that common target genes of multiple miRNAs can be easily recognized in this miRNA–target mRNA table.
Figure 2.
Sample output picture from the target–function–expression module.
Input of multiple genes can be used to find the regulatory miRNAs for the given set of genes. If genes belonging to a specific biological pathway are provided as an input, miRNAs with multiple target genes within the pathway of interest may be identified. Similar approach can be applied to find the regulatory miRNAs for gene clusters obtained from mRNA expression profiling.
Functional enrichment analysis of target genes can be performed in three categories—GO, pathway and disease terms. Simple hypergeometric test of over-representation in each term was carried out for all terms in GO, pathway and disease classification systems. The output page summarizes the significant nodes for a given P-value, which can be sorted according to various criteria. Our pathway analysis includes the KEGG, GenMAPP and BioCarta pathway databases. Disease classification of the Ingenuity systems Inc. was used to test disease implications. Since all calculations are pre-computed and stored in the database, the search for miRNAs whose target genes are statistically enriched in specific terms is also possible. A separate module of ‘miRNA with inferred function’ is provided to look for the miRNAs with inferred functions in all three functional categories. Our web implementation supports any node id in the GO classification and all pathways. As for the disease search, we support 29 representative terms only.
Expression correlation analysis of target genes gives a table of correlation coefficients between miRNA and target mRNA as well as miRNA and target protein if the data are available. Reciprocal expression pattern is expected for genuine targets and the pairs of high correlation between miRNA and apparent non-targets may indicate indirect targeting. Target genes can be sorted according to the correlation coefficients in descending or ascending order. Link to detailed information on target binding and correlated expression pattern is also provided for each miRNA–mRNA pair.
MicroRNA expression profiling module
The purpose of miRNA expression profiling module is to visualize miRNA expression and to obtain information on differential regulation in various situations. We performed 106 comparisons from 12 GEO experiments. Simple query of miRNA, tissue/organ name, or disease yields a list of relevant comparison studies. Once miRNA and comparison study are specified, expression pattern of miRNA across the samples in the study can be displayed as a bar or box plot.
Searches other than miRNA allow the user to access the list of differentially regulated miRNAs in various situations. Comparison studies were classified into subgroups according to its purpose. Current implementation supports searches for differentially regulated miRNAs in 24 tissues/organs and 28 cell types. Comparing expression conditions consists of four types of cancer versus normal, cancer versus cancer, chemical treatment and others. For example, current dataset of comparing cancer versus normal tissues supports seven tissues including bladder, breast, colon, eye, kidney, lung and uterus.
ACKNOWLEDGEMENTS
This work was supported by a grant (No. 20070401034010) from BioGreen 21 Program of the Korean Rural Development Administration and by the Korean Ministry of Science and Technology through the bioinformatics research program. B.K. is grateful for the BK21 research fellowship from the Ministry of Education and Human Resources Development. Funding to pay the Open Access publication charges for this article was provided by the Korean Rural Development Administration.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 3.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilsen T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl. 1):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 8.Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 9.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Kuhn R.M., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sethupathy P., Corda B., Hatzigeorgiou A.G. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human MicroRNA targets. PLoS Biol. 2004;2:E363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shahi P., Loukianiouk S., Bohne-Lang A., Kenzelmann M., Kuffer S., Maertens S., Eils R., Grone H.J., Gretz N., et al. Argonaute – a database for gene regulation by mammalian microRNAs. Nucleic Acids Res. 2006;34:D115–D118. doi: 10.1093/nar/gkj093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu P.W., Huang H.D., Hsu S.D., Lin L.Z., Tsou A.P., Tseng C.P., Stadler P.F., Washietl S., Hofacker I.L. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–D139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Megraw M., Sethupathy P., Corda B., Hatzigeorgiou A.G. miRGen: a database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007;35:D149–D155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiriakidou M., Nelson P.T., Kouranov A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahlquist K.D., Salomonis N., Vranizan K., Lawlor S.C., Conklin B.R. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 22.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 23.Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B., Barrette T.R., Anstet M.J., Kincead-Beal C., et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camon E., Magrane M., Barrell D., Lee V., Dimmer E., Maslen J., Binns D., Harte N., Lopez R., et al. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung H.J., Park C.H., Han M.R., Lee S., Ohn J.H., Kim J., Kim J., Kim J.H. ArrayXPath II: mapping and visualizing microarray gene-expression data with biomedical ontologies and integrated biological pathway resources using Scalable Vector Graphics. Nucleic Acids Res. 2005;33:W621–W626. doi: 10.1093/nar/gki450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Babak T., Zhang W., Morris Q., Blencowe B.J., Hughes T.R. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W., Morris Q.D., Chang R., Shai O., Bakowski M.A., Mitsakakis N., Mohammad N., Robinson M.D., Zirngibl R., et al. The functional landscape of mouse gene expression. J. Biol. 2004;3:21. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., et al. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 29.Ramaswamy S., Tamayo P., Rifkin R., Mukherjee S., Yeang C.H., Angelo M., Ladd C., Reich M., Latulippe E., et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc. Natl Acad. Sci. USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]