Abstract
The pathogen–host interaction database (PHI-base) is a web-accessible database that catalogues experimentally verified pathogenicity, virulence and effector genes from bacterial, fungal and Oomycete pathogens, which infect human, animal, plant, insect, fish and fungal hosts. Plant endophytes are also included. PHI-base is therefore an invaluable resource for the discovery of genes in medically and agronomically important pathogens, which may be potential targets for chemical intervention. The database is freely accessible to both academic and non-academic users. This publication describes recent additions to the database and both current and future applications. The number of fields that characterize PHI-base entries has almost doubled. Important additional fields deal with new experimental methods, strain information, pathogenicity islands and external references that link the database to external resources, for example, gene ontology terms and Locus IDs. Another important addition is the inclusion of anti-infectives and their target genes that makes it possible to predict the compounds, that may interact with newly identified virulence factors. In parallel, the curation process has been improved and now involves several external experts. On the technical side, several new search tools have been provided and the database is also now distributed in XML format. PHI-base is available at: http://www.phi-base.org/.
INTRODUCTION
Many microbes have the potential to cause disease. The pathogen–host interaction database (PHI-base), established in 2005, contains expertly curated molecular and biological information on genes proven to affect the outcome of a pathogen–host interaction. PHI-base catalogues the genes confirmed through gene disruption and/or transcript level alteration experiments to be required for the disease causing ability of a microbe. These genes are termed pathogenicity genes if the effect on the phenotype is qualitative or virulence/aggressiveness genes if the effect is quantitative. An additional category of genes included in PHI-base are effector genes, which either activate or suppress plant defence responses. Four reasons have provided the motivation to improve further the functionality of PHI-base: (i) In the post-genomics era the efficiency of both forward and reverse genetics analyses to determine the function of the encoded gene product(s) has greatly accelerated. (ii) There is also intense interest in comparative genomics, to identify both functionally homologous genes and species-unique genes. (iii) Full genomic sequence is becoming available for additional strains of a single species. This can be used to reveal subtle sequence differences, which may confer altered interaction phenotypes. (iv) Finally, and most importantly, researchers require free and easy access to different types of interaction information. This permits both interactive and custom analyses of genes of interest to be made, which facilitates hypothesis generation and knowledge discovery.
The original article on PHI-base was published in the NAR database issue in 2006 (1). A second article reviewed in detail the biological contents of PHI-base version 2.1 and described the emerging unique features associated with plant pathogenesis (2). The PHI-base website now receives about 5000 hits per quarter (excluding search engines and internal users). The database has been cited for various purposes. For example, Jeon and colleagues (3) used PHI-base to annotate the results of an extensive forward genetics experiment which had identified >200 mutants of the economically important rice blast fungal pathogen Magnaporthe grisea with reduced ability to cause disease on rice leaves. DiGuistini and colleagues (4) annotated a population of ESTs arising from a Mountain Pine Beetle-associated fungal pathogen even though no entries for a pathogen with this mode of pathogenesis exist in PHI-base. Sexton and Howlett (5) used the contents of version 2.3 as the backbone for a comprehensive review of the common and unique requirements of animal and plant pathogenesis during initial infection, colonization and sporulation on the host.
Philippi and Köhler (6) describe 10 essential quality criteria for biological databases that should be met as a prerequisite to the publication of any scientific database. The PHI-base changes and updates described here address all these quality criteria as well as the changes that were necessary to improve the breadth and depth of data captured in PHI-base and to provide improved user access.
NEW FEATURES AND DETAIL
A broader range of pathogenicity and effector genes
Version 2.1 exclusively contained information on fungal and Oomycete pathogens. PHI-base 3.0 now also includes bacterial pathogens and plant endophytes. Currently the database contains 786 pathogenicity genes from fungi, 27 pathogenicity genes from Oomycetes and 137 from bacteria. Most bacterial genes are from plant pathogens, but some human pathogens such as Vibrio cholerae are also captured. For many pathogen species the effects of single, double and triple gene mutations in a single strain infecting multiple host species are provided and these entries are interlinked.
A second important addition requested by users was the inclusion of gene homologues, which when made non-functional did not alter the disease causing ability of the organism. Inclusion of these results (pathogenicity not affected—a total of 211 genes/288 interactions) is very useful for comparative genomics analyses when, for example, investigating the function of putative conserved pathways and/or proteins.
Many new techniques for investigating gene product function have become more widely adopted, for example, gene silencing (7) and transient gene expression (8). PHI-base now incorporates genes that were identified by these new methods. For example, for entries PHI:758 and PHI:759, seven different sources of weaker experimental evidence were used to verify the identification of the first effector genes for the non-culturable barley powdery mildew fungus (Blumeria graminis f. sp. hordei).
Anti-infectives and their pathogen target site(s)
Another important addition to PHI-base was the inclusion of verified targets of known bioactive compounds (anti-infectives) which either kill pathogens or arrest pathogen growth/development. The genes encoding these targets are often not pathogenicity genes in the strict sense, but are instead considered to be ‘essential-for-life’ genes. Using standard techniques such as deletion/disruption experiments, signature-tagged mutagenesis or promoter alterations, a lethal/highly debilitating phenotype is frequently observed. These genes have very important applications. For example, sequence similarity comparisons make it possible to infer across species which existing bioactive compounds could also be used as anti-infectives against newly emerging pathogens and/or strains. To support this type of comparative analysis, the fungicides and target genes compiled by the fungicide resistance action committee (FRAC, http://www.frac.info) have been included in PHI-base. We have also started to include anti-infectives for human pathogens in PHI-base cited by the British National Formulary (http://www.bnf.org/bnf/search.htm).
Provision of greater detail
To capture in full the characteristics and the experimental evidence supporting the pathogenicity genes and anti-infective target sites, the number of new fields describing each PHI-base entry has almost doubled since the 2.1 release. In addition, some obsolete fields were removed and all existing entries revised. Besides the data fields required to capture the chemical characteristics of the new anti-infectives, pathogenicity islands, enzyme commission (EC) numbers, UniProt database identifiers, genome locations and deep links to the referenced literature using digital object identifiers (DOIs), have been added, where appropriate. Strain and subspecies information is now included with a distinction made between the sequenced strain of the organism and the strain under study in the PHI-base entry. In addition, several new structured fields using controlled vocabularies have now been created. A specific interest group, the Plant-Associated Microbe Gene Ontology Interest Group (PAMGO), was set up in 2004 to ensure that specific gene ontology (GO) terms are developed for plant–microbe interactions (7). Since then the PAMGO consortium, in collaboration with the gene ontology consortium has approved and released 472 new terms for describing gene products involved in microbial–host interactions (http://pamgo.vbi.vt.edu/index.php). Free text function annotations have been replaced with adequate gene ontology (GO and PAMGO) terms.
New database entries
The version 3.0 release now has 950 manually curated entries, whereas the 2.1 version contained about 427 entries. The new contents are summarized in Tables 1 and 2.
Table 1.
Summary of the number of pathogen and host species and genes within version 3.0 of PHI-base
| Host/Entry type | Vertebratea | Planta | Fungal | Insect |
|---|---|---|---|---|
| Host species | 8 | 51 | 3 | 3 |
| Genes in totalb | 339 | 588 | 3 | 8 |
| Pathogenicity genes | 50 | 97 | 0 | 0 |
| Virulence genes | 243 | 241 | 3 | 6 |
| Effector genes | 0 | 64 | 0 | 0 |
| Anti-infective targetc | 5 | 19 | 0 | 0 |
aTotal entry numbers for each of the top three vertebrate and plant-attacking species are 353 and 288, respectively (the same gene can be tested for function on different hosts from different kingdoms).
bSome genes were tested on more than one host, some genes have no specific host.
cThirteen anti-infective targets are broad-range and have no specific host associated.
Table 2.
Summary of the number of pathogen species and genes within PHI-base version 3.0
| Pathogen/Entry type | Fungal | Oomycete | Bacterial |
|---|---|---|---|
| Pathogen species | 72 | 5 | 18 |
| Genes in total | 786 | 27 | 137 |
| Pathogenicity genes | 147 | 0 | 0 |
| Virulence genes | 422 | 70 | 3 |
| Effector genes | 11 | 21 | 38 |
| Anti-infective target | 30 | 0 | 7 |
TECHNICAL DEVELOPMENTS, CURATION AND OUTREACH
Data curation and release management
To ensure a consistently high quality for all entries in PHI-base, a closer interaction with the global user community was established. This also improved the information content most relevant to the end users. PHI-base version 2.1 was developed by training scientists at Rothamsted to curate data and to find the appropriate articles via text mining. Since early 2006, effective communication routes have been provided to enable users to report errors in the database and to suggest new entries and new features to the database. Several external domain experts for specific pathogen species now actively contribute to the database by suggesting new entries and by revising existing entries as part of the beta-testing phase. We always welcome the contribution and participation of new experts for organisms not yet included in PHI-base.
Currently, most species experts contribute to PHI-base, by referring the trained curators to relevant publications, which describe genes that are valid for inclusion in PHI-base. These genes are then added to the database by a curator at Rothamsted Research. Each new entry is subsequently checked and approved by the species expert and a second local expert curator. However, some species experts directly curate new entries. These are checked for completeness and the correct entry format by the PHI-base team before being uploaded into the database. User documentation is provided on the search interfaces and the content.
To deal effectively with the larger user and contributor community, the release management process has been further formalized. The first step of creating a new release is to supplement the manually curated content of PHI-base with imported information, for example, sequences and GO terms from EMBL. This is followed by a beta-testing phase of several weeks in which newly added entries and revised fields are checked by the Rothamsted curators and external domain experts. At the same time, the technical functionality of the database is tested using a set of standard queries.
Data exchange and interfaces
For users wishing to embed PHI-base data in other applications, the terms of use have been relaxed. Redistribution and integration of PHI-base into other applications is now permitted, be it in part or as a whole, as long as the redistributing database operates under the same licencing conditions and that the PHI-base source is clearly displayed within each entry.
The search interface has been extended. All entries can be searched using a full text search facility. Alternatively, the new advanced query interface allows users to exploit the structured way in which PHI-base stores data using appropriate searches. Three multi-selection fields have been created that allow for the specific retrieval of information on the phenotypes obtained, the experimental evidence provided and gene products that are known targets for certain groups of anti-infectives.
The complete database or the results from specific searches, including all the details, which are provided in the online version of PHI-base, can now be downloaded using XML. As a consequence, it is possible to load directly either PHI-base or search results from PHI-base into the data integration system ONDEX (8) and then into other external applications, for example, the KEGG pathway databases. In version 2.1, only a Fasta dump was provided of the sequence information in PHI-base.
APPLICATIONS OF PHI-BASE
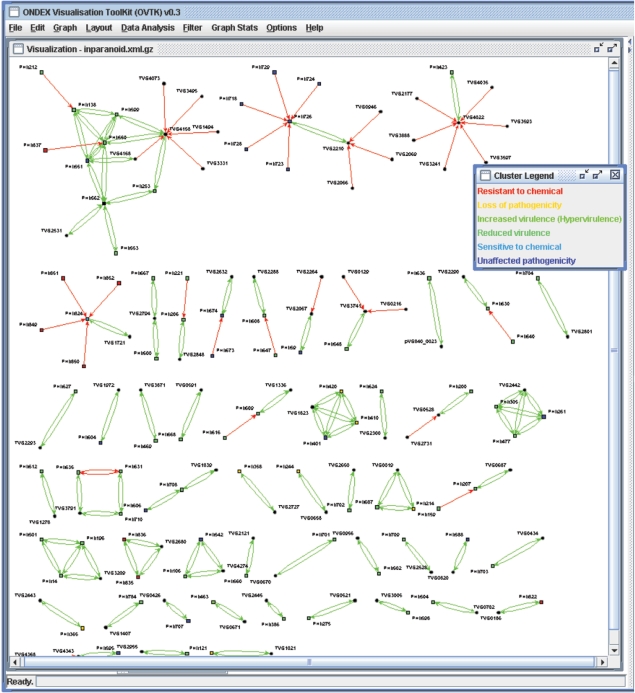
The most common application of PHI-base is to use the database for the identification of pathogenicity genes arising either from comparative genomics analyses (3) or from forward genetics analyses in a different organism (2). However, the PHI-base data is also increasingly being used to annotate newly sequenced genomes and EST projects. For example, using the ONDEX system (8), the genome of the fish pathogen Vibrio salmonicida was annotated effectively (Figure 1). To identify putative virulence factors in this newly sequenced organism, the following steps were done within the data integration system ONDEX: (i) XML format of PHI-base uploaded; (ii) homologues identified using an e-value of <10−6; (iii) BLAST hits further filtered so that only genes which aligned across more than 50% of their length remained and (iv) the results were loaded into the graph analysis component of the ONDEX system (Figure 1). This analysis identified 50 putative virulence factors in V. salmonicida.
Figure 1.
Predicted virulence genes for the newly sequenced fish pathogen V. salmonicida (Vs). Vs genes are displayed as circles, PHI-base genes are displayed as rectangles and are colour coded to indicate type. The arcs between the genes represent sequence–homology relationships. The visualization tool has placed ∼50 putative Vs virulence genes in V. salmonicida into distinct clusters of PHI-base entries.
When investigating the function and evolution of specific pathogenicity factors, the inclusion of natural and induced gene sequence variants is proving to be particularly informative (for example, PHI:7 and PHI:71). Also when exploring the population dynamics of a species either locally or globally, rather than choose a set of neutral gene markers (9), it is possible to select sets of pathogenicity and/or effector genes of known biological importance. Finally, in the post-genomics era, vast quantities of transcriptomics and proteomics data are available for many pathogenic organisms and their hosts. By identifying which genes are coordinately expressed with PHI-base entries and/or their homologues this should greatly assist in the characterization and experimental prioritization of genes with currently no functional annotation.
UPCOMING DEVELOPMENTS
In the next version of PHI-base the host target of pathogen effectors will be included. For several plant bacterial pathogens these key components present in both susceptible and resistant hosts have recently been identified [reviewed Ref. (10)]. The next logical step will be the inclusion of the many known components of the functionally effective host defence signalling networks. Currently, most of the information on the latter is contained within single organism databases and most entries provide only general information.
Although this release of PHI-base contains some bacterial pathogen entries, the database is still heavily biased towards fungal and Oomycete pathogens. More species experts for plant, human and animal bacterial pathogens will be involved to assist with these entries. Subsequently, viral pathogens will also be included. To improve the value of PHI-base for comparative purposes additional beneficial symbionts, will be included such as the plant invading nitrogen-fixing bacteria and nutrient-acquiring mycorrhizal fungi (11).
To improve the curation process and involve a wider community of species experts, two types of web-based interfaces will be developed. One will enable users and species experts to suggest new entries for inclusion to the database and will only capture basic information. The second will be an expert curation interface that will be developed to enable trained curators to enter all required detail in a fast and efficient way.
To enable programmers and users of workflow management tools such as Taverna (12) to use PHI-base, we plan to provide Web Service interfaces that can be accessed computationally which makes it possible to include PHI-base into complex data analysis and integration tasks.
The inclusion of key GO/PAMGO terms within publications and the corresponding MEDLINE abstracts would greatly improve the identification of papers which describe new pathogenicity determinants. For example, the parent term GO:0044403—‘symbiosis, encompassing mutualism through parasitism’ currently contains 956 entries in MEDLINE. However, the majority of papers are still published without such systematic annotations.
DATABASE ACCESS AND FEEDBACK
PHI-base can be freely accessed at http://www.phi-base.org/. User support can be obtained from this email: phi-base@bbsrc.ac.uk. Please use the same email address if you wish to provide new data for inclusion in PHI-base, are an interested expert willing to assist with curation or can provide suggestions for improvements.
ACKNOWLEDGEMENTS
The authors would like to thank the FRAC team for allowing inclusion of their data on fungicides and the PAMGO group for compiling a useful ontology on microbe–host interactions. We are grateful to all species experts who contributed additional database entries from their field of expertize, namely Roland Krause (Max-Planck Institute for Molecular Genetics, Germany), Jason Rudd (Rothamsted Research, UK), Chris Ridout (John Innes Centre, UK), Jan Schirawski (Max-Planck Institute for Terrestrial Microbiology, Marburg, Germany), Paul Tudzynski (Institut für Botanik, Münster, Germany), Jan van Kan (Wageningen University, Laboratory of Phytopathology, The Netherlands), Barry Scott (Institute of Molecular Biosciences, Massey University, New Zealand) and Alexander Idnurm (Duke University Medical Center, Durham, NC, USA). We would also like to thank Jan Taubert (Rothamsted Research, UK) for the ONDEX analysis as an example for possible applications of PHI-base. Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council (BBSRC). Sabrina Holland would like to thank the EU for funding through the Leonardo scholarship scheme. Funding to pay the Open Access publication charges for this article was provided by the BBSRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Winnenburg R., Baldwin T.K., Urban M., Rawlings C., Köhler J., Hammond-Kosack K.E. PHI-base: a new database for pathogen host interactions. Nucleic Acids Res. 2006;34(Database issue):D459–D464. doi: 10.1093/nar/gkj047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin T.K., Winnenburg R., Urban M., Rawlings C., Koehler J., Hammond-Kosack K.E. The pathogen-host interactions database (PHI-base) provides insights into generic and novel themes of pathogenicity. Mol. Plant Microbe Interact. 2006;19:1451–1462. doi: 10.1094/MPMI-19-1451. [DOI] [PubMed] [Google Scholar]
- 3.Jeon J., Park S.Y., Chi M.H., Choi J., Park J., Rho H.S., Kim S., Goh J., Yoo S., et al. Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat. Genet. 2007;39:561–565. doi: 10.1038/ng2002. [DOI] [PubMed] [Google Scholar]
- 4.DiGuistini S., Ralph S.G., Lim Y.W., Holt R., Jones S., Bohlmann J., Breuil C. Generation and annotation of lodgepole pine and oleoresin-induced expressed sequences from the blue-stain fungus Ophiostoma clavigerum, a mountain pine beetle-associated pathogen. FEMS Microbiol. Lett. 2007;267:151–158. doi: 10.1111/j.1574-6968.2006.00565.x. [DOI] [PubMed] [Google Scholar]
- 5.Sexton A.C., Howlett B.J. Parallels in fungal pathogenesis on plant and animal hosts. Eukaryot. Cell. 2006;5:1941–1949. doi: 10.1128/EC.00277-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philippi S., Köhler J. Addressing the problems with life-science databases for traditional uses and systems biology. Nat. Rev. Genet. 2006;7:482–488. doi: 10.1038/nrg1872. [DOI] [PubMed] [Google Scholar]
- 7.Lindeberg M., Stavrinides J., Chang J.H., Alfano J.R., Collmer A., Dangl J.L., Greenberg J.T., Mansfield J.W., Guttman D.S. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol. Plant Microbe Interact. 2005;18:275–282. doi: 10.1094/MPMI-18-0275. [DOI] [PubMed] [Google Scholar]
- 8.Köhler J., Baumbach J., Taubert J., Specht M., Skusa A., Ruegg A., Rawlings C., Verrier P., Philippi S. Graph-based analysis and visualization of experimental results with ONDEX. Bioinformatics. 2006;22:1383–1390. doi: 10.1093/bioinformatics/btl081. [DOI] [PubMed] [Google Scholar]
- 9.O'Donnell K., Kistler C.H., Tacke B.K., Casper H.H. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl Acad. Sci. USA. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 11.Tunlid A., Talbot N.J. Genomics of parasitic and symbiotic fungi. Curr. Opin. Microbiol. 2002;5:513–519. doi: 10.1016/s1369-5274(02)00355-7. [DOI] [PubMed] [Google Scholar]
- 12.Oinn T., Addis M., Ferris J., Marvin D., Senger M., Greenwood M., Carver T., Glover K., Pocock M.R., et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004;20:3045–3054. doi: 10.1093/bioinformatics/bth361. [DOI] [PubMed] [Google Scholar]