Abstract
The multitude of bacterial genome sequences being determined has generated new requirements regarding the development of databases and graphical interfaces: these are needed to organize and retrieve biological information from the comparison of large sets of genomes. GenoList (http://genolist.pasteur.fr/GenoList) is an integrated environment dedicated to querying and analyzing genome data from bacterial species. GenoList inherits from the SubtiList database and web server, the reference data resource for the Bacillus subtilis genome. The data model was extended to hold information about relationships between genomes (e.g. protein families). The web user interface was designed to primarily take into account biologists’ needs and modes of operation. Along with standard query and browsing capabilities, comparative genomics facilities are available, including subtractive proteome analysis. One key feature is the integration of the many tools accessible in the environment. As an example, it is straightforward to identify the genes that are specific to a group of bacteria, export them as a tab-separated list, get their protein sequences and run a multiple alignment on a subset of these sequences.
INTRODUCTION
Since the publication of the first bacterial genome sequence (1), strategies and technologies for whole genome sequencing have continuously improved. As a consequence, there are >520 publicly available bacterial genomes at the time of writing (http://www.genomesonline.org/). This wealth of information has generated new requirements for efficient and specialized data-mining tools (2,3). Indeed comparative genomics entails the development of novel methods, databases and graphical interfaces to organize, browse and retrieve biological information from the analysis of numerous genome sequences (4). International sequence databanks have faced new challenges to manage this profusion of data, and partially met them by creating novel resources (5–7). In addition, a number of specialized databases dedicated to the analysis of microbial genomes have been established (8–12).
Upon publication of the complete genome sequence of the Gram-positive bacterium Bacillus subtilis (13), an original database model was defined along with an innovative web interface, allowing biologists to efficiently browse and query the related information (14,15). The SubtiList database was derived from previous developments dedicated to Escherichia coli DNA sequence contigs, going back to the pregenomic era (16). Later on, the genome sequences of two strains of the same species were made available—those of Helicobacter pylori (17,18)—and the SubtiList model was extended to hold the two sequences and annotation sets simultaneously (19). Numerous individual databases based on the same model were set up since then [http://genolist.pasteur.fr and (20)].
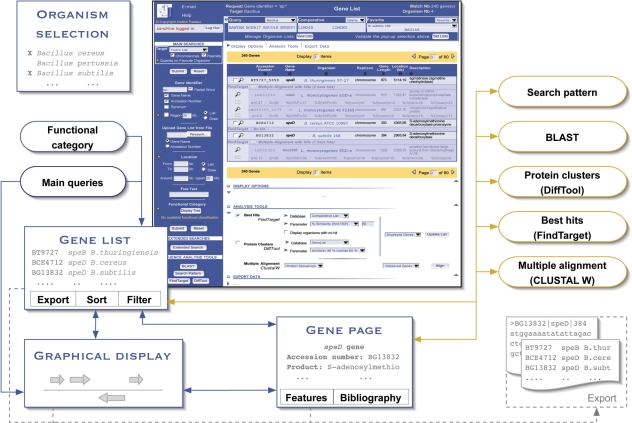
Building upon these successive accomplishments, the GenoList multigenome environment was created: it consists of a relational database and a dynamic web application, aimed to take advantage of the multitude of published bacterial genomes to perform data analysis in a comparative genomics context. The development of GenoList was motivated by four key targets: (i) to organize microbial genome information in a specialized relational data schema, making it possible to implement complex relationships and subsequent queries; (ii) to supplement public genome data with original information, such as expertly curated annotations and comparative genomics data; (iii) to provide biologists with specific query and exploratory facilities conceived in an end-user-oriented fashion and (iv) to integrate the data content and the many search and analysis tools to seamlessly link up series of queries/responses (Figure 1). GenoList thus provides the user with an integrated environment that generates an added value compared to the initial data, both through the inclusion of original supplementary information and by enabling innovative functionalities for data study and extraction. GenoList is accessible at the URL http://genolist.pasteur.fr/GenoList.
Figure 1.
Data and functionality integration in the GenoList environment. The first step consists in selecting species of interest (‘Organism selection’ panel). The central component of the application is the ‘Gene list’ panel (shown in the screen capture). Search outputs are presented there (‘Main queries’ and ‘Functional category’—blue rounded boxes), and results of sequence analysis tools (‘Search pattern’, ‘BLAST’, ‘DiffTool’ and ‘FindTarget’—yellow rounded boxes), which are displayed using specific layouts (e.g. BLAST reports), can also be converted to standard ‘Gene lists’. This panel acts as a hub since it gives access to many additional features (‘Gene page’, ‘Graphical display’ and ‘Export’), and it allows the user to bounce back to the sequence analysis tools from either one single gene or a subselection of the gene list. The screen capture illustrates the visualization of best hits of selected genes in a standard ‘Gene list’.
Data Content and Organization
Internal data structure
The relational data model that underlies the GenoList database comprises >80 tables, implemented in SQL using the Sybase ASE (Sybase Inc.) database management system. It was designed with three main ideas in mind: being generic enough to enable the modeling of any kind of features found in microbial genomes; allowing the most frequent queries performed by biologists to run flawlessly; establishing relationships between genomes through comparative genomics data. As an example, replicon sequences and associated genomic objects were artificially split into short fragments of predefined length, to avoid performance issues during query and update operations of very large genomic sequences (this obviously remains transparent to the end user). Similarly, a clear separation was created between ‘physical’ (genome coordinates) and functional genomic objects, thus ensuring a high level of standardization in the treatment of these items. The gene and protein concepts were given a structured definition including several levels of representation (coding sequences, RNA genes, protein products, complexes, etc.). Reliable identifiers were defined for the main objects of the database, both external ones—public accession numbers, gene names—and internal ones—specific nomenclature used in GenoList for internal consistency purposes. The data model makes it possible to create relationships between genomic objects, both inter- and intra-organisms. Especially, protein families can be represented in two different ways: either transitive associations (i.e. partitions obtained by a classification method) or non-symmetrical relationships used for the connection of one specific gene with a set of other genes (e.g. BLAST reports, ‘best hits’—see below).
Genome data sources
The current data release of GenoList (R2.0) contains information from 103 bacterial genomes. They were selected among publicly available genome sequences in accordance with interests shown by the first users of GenoList. As a result, they represented a coherent subset of taxa, which was favorable for load testing of the application, and will be extended in upcoming releases. Genome sequences and annotations were retrieved and parsed from original GenBank entries (21) and integrated into the database using an ‘Extract-Transform-Load’ (ETL) pipeline. This pipeline implemented in Perl (BioPerl parser framework) was driven by shell scripts that managed the automatic download of source databanks from external ftp servers, the parsing of the flat files, and the preparation of load files for database feeding. Genomes were organized in the database according to the taxonomic classification of the NCBI (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html).
Due to the heterogeneous quality standard of genome annotations in public data collections and due to the lack of regular updates, GenBank entries were replaced in a few cases by re-annotated genome sequences, exported from the databases available in the first generation of mono-genome GenoList web servers (http://genolist.pasteur.fr/). These include the genomes of B. subtilis (15), H. pylori (19) and Mycobacterium tuberculosis (22). It is indeed essential that genome data from reference species remain as current as possible (23). Scientific literature was thus attached to genes from the aforementioned species, together with relevance values selected from a controlled vocabulary and indicating the underlying principle of each gene-reference association. In the current data release (R2.0), ca. 10 000 gene annotations were expertly curated. Efforts will be pursued to offer in GenoList high-quality genome annotations, which take into account the continual improvement of bioinformatics techniques for gene and function prediction, and the unceasing production of novel experimental facts. In particular, the integration of data from updated repositories such as Genome Reviews (6) and RefSeq (7) will be considered.
In-house precomputed data
In addition to the innovative graphical user interface described below, GenoList builds upon GenBank and re-annotated genome data to generate further information about genome and protein sequences. For example, transitive protein families were precomputed using the DiffTool algorithm (24), which was inspired from the well-recognized HOBACGEN specifications (25): BLASTP searches (26) were run using each protein in GenoList as a query sequence against all other individual proteins; the proteins that shared at least x% of similarity over an amino acid overlap of length y were clustered together, using a transitive closure rule. The clustering step was performed several times with distinct parameters x and y to produce a number of family sets. The parameter y was generally large enough (ca. 80%) to avoid the clustering of multidomain proteins. Protein families were stored in dedicated tables of the database so as to be queried using the relevant forms of the user interface (see below). The database model makes it possible to store other types of protein families computed externally (e.g. the Clusters of Orthologous Groups of proteins (27)): such protein clusters will be integrated in upcoming releases of GenoList. Using a procedure similar to DiffTool (lacking the clustering step) called FindTarget (28), a systematic calculation of BLASTP ‘best hits’ was performed for all proteins against all individual proteomes present in GenoList. These best hits were stored in the database in order to be used at the user interface level, both on a gene-by-gene basis and in global subtractive proteome analysis (see below). Finally, as introduced in the SubtiList database a decade ago (15), BLASTP reports (26) were produced by running each protein stored in GenoList as a query sequence against the UniProt databank (29). These reports—meant to be updated on a regular basis—were kept as flat files linked to the database for immediate access through the user interface.
User Interface
A graphical user interface was designed to access the GenoList database on the web. Its development was guided by the querying rationale of biologists. Following discussions with current and future users, the typical and most frequent means of access to the data were identified, and an intuitive user-friendly graphical interface was built to fulfill these requirements. It allows one to browse and visualize data in various ways, especially through a rapid access to comparative analysis.
The web application was written in the Java language, using the object-oriented framework WebObjects (Apple Inc.). WebObjects is both an integrated development environment and a web application server, based on a three-tier architecture with powerful data connectivity features and robust business logic for creating and deploying scalable and easy-to-maintain web applications.
Organism selection
Upon accessing the web application, the user may either log on (after having registered) or directly select the organisms he will work with. Registering and login are optional. However, this is the recommended mode of use of the environment, as it allows the user to record a customized configuration that will be automatically recalled at the beginning of every working session. This includes the definition of organism lists delineating species of interest. Organisms whose genome is stored in the database can be selected to build three types of lists: the ‘Query’ type is used as the default selection of species targeted by main searches; the ‘Comparative’ type is used in various comparative analysis (see below); the ‘Favorite’ type consists of one single organism, used by default for every action that requires a single selection. Several query, comparative and favorite lists can be defined and permanently stored so that they remain accessible to registered users upon return to the database. To create new lists, either an alphabetical view or a taxonomic representation can be used: the latter allows the user to easily select a whole range of species at a given taxonomic level. Once organism lists are specified, it is straightforward to shift working lists in the course of a session, thanks to pop-up menus directly accessible in the main application page (Figure 1). Thus, by defining organism lists, one can use the GenoList database while focusing on one or several species, as if only the latter were present in the database.
Overall layout and query features
The front page of the application is organized in two components: the left-hand area contains text fields and menus required to perform queries on the database, whereas the right-hand part holds results generated from searches and analysis (Figure 1). Similarly to the web interface of SubtiList (15), the most frequent types of request (e.g. gene identifier, genome region—either around a gene or between coordinates, etc.) are directly available from the main page. A pop-up menu allows the user to rapidly select the set of organisms to be considered as the query target: either one of the predefined list of species or all genomes present in GenoList. Advanced queries and sequence analysis require the display of specialized forms in the right-hand section (see below).
Gene lists and gene pages
Most queries generate gene lists, which are a central component of the environment (Figure 1). Gene lists observe a highly customizable layout that allows the user to choose which kinds of information (columns) he wishes to see. In addition, taking advantage of the precomputed comparative data (see above), the application provides options to display gene best hits processed by the FindTarget algorithm (28), and/or protein families the genes belong to according to the DiffTool algorithm (24) (Figure 1). By default, these options use the selected ‘Comparative’ list of organisms to retrieve best hits and families; however, this parameter remains user adjustable, just like the sequence identity/similarity threshold to be used. Subsequently, the corresponding individual proteins can be aligned using pairwise or multiple sequence alignment methods [e.g. CLUSTAL W (30)]. Gene lists can also lead to customizable graphical representations of genome regions (when applicable), and provide means to export data in various formats (tab-separated gene information, DNA/RNA/protein sequences, etc.). Most importantly, gene lists constitute a hub between searches and results: they are intended to display query and analysis outputs (whether it be simple gene lists, genome regions, gene families, etc.), and at the same time they make it possible to launch several data handling and analysis tools (e.g. data export, display of comparative genomics results, multiple sequence alignment, etc.). This way, the application can be used through loops of queries responses, which happen to be an efficient strategy to explore data (Figure 1).
A checkbox located beside each gene in a list allows the user to define a subselection of genes on which the operations described above can be performed. Also, each individual gene can be viewed in a separate window that gives further information about the gene of interest. The gene page is organized in several tabs: these include functional information and protein features (when available), a graphical representation of the genomic neighborhood, comparative genomics data (both precalculated and computable with user-defined parameters), precomputed up-to-date BLASTP reports against a non-redundant protein databank, cross references with external data collections (e.g. GenBank (21), Swiss-Prot (29), InterPro (31), etc.), bibliographical references (when available) and sequence information. Without leaving the gene page, the user can shift the gene displayed by navigating in the current gene list or in the current replicon.
Sequence analysis functionalities
Sequence analysis tools, such as the BLAST databank scanning algorithm, and an ambiguous DNA/protein pattern search utility, are directly accessible from the web application. Options to these analyses were devised to capitalize on the integration of the tools within the database environment. For instance, it is possible to define which DNA pattern results should be displayed depending on their location with respect to gene coordinates (start and stop codons, intergenic regions, etc.). The FindTarget and DiffTool subtractive proteomics algorithms were also integrated into the GenoList interface. FindTarget (28) makes it possible to identify genes from a given organism (‘Query Genome’—the selected ‘Favorite’ organism) that are specifically present in a set of species (‘Reference Genome(s)’—by default the ‘Query’ list), and, optionally, absent in another set of species (‘Exclusion Genome(s)’—by default the ‘Comparative’ list). The reference and exclusion genomes, as well as the sequence identity/similarity thresholds used by the algorithm, can be set on the fly. Similarly, DiffTool (24) allows the user to search for protein clusters that contain proteins from the selected ‘Reference Genome(s)’ and, optionally, no protein from the selected ‘Exclusion Genome(s)’. Several sets of protein families were stored in GenoList (differing as a function of the clustering parameters used—see the ‘In-house precomputed data’ section) and can be selected at the time of the search. Such differential studies on whole proteomes allow the identification of specific genes shared by a set of species as compared to another set: this is a typical analysis performed when comparing pathogenic microorganisms with non-pathogenic-related strains.
Conclusion and Perspectives
The rationale that prevails at the development of the GenoList system is the creation of a specialized environment for biologists. GenoList aims to act as an information hub that connects logically structured high-quality data and relevant exploratory tools, accessible through graphical user interfaces designed to respond to scientists’ needs; this ideal objective obviously asks for a continuous enhancement of the information stored in the database (e.g. only a few genome annotations were expertly curated to date), and for further innovative functionalities that will allow users to mine the data in novel ways. An added value is thus generated by the synergy between the individual components of an integrated software environment: this contributes to knowledge discovery through fine data exploration, which is guided by suitable user-driven visual representations and human–machine interfaces.
Future developments of the GenoList database will follow several paths. Further complete bacterial genomes will be progressively entered, although exhaustiveness is not a necessary objective: indeed including too many genomes in the same database may end up hindering the initial goal of the environment, thus priority will be given to species of interest requested by GenoList users. Conversely, it should be noted that the facility provided by user-defined lists of organisms, allowing one to easily restrict searches and analysis to selected species only, may alleviate the genome-crowding issue. The inclusion of unfinished genomes will be considered since the data model was designed from the beginning so that it could house such data. An extension of GenoList to eukaryotic species has recently been implemented in order to provide the CandidaDB database (32), dedicated to Candida albicans and related species, with a multigenome dimension (33).
Additional innovative tools are to be implemented in order to maximize the integration of multiple genomes into a single framework, along with comparative relationship information. Multigenome queries using concepts such as synteny or phylogenetic profiles will be devised (34). Moreover, new modules for complex data searches and elaborate sequence retrieval are in the planning stages. Finally, linking GenoList to other resources will enhance data-mining functionalities, taking advantage of cross queries made possible between heterogeneous databases (35). In particular, integration of annotation data and functional genomics information is a major issue in tackling a systems-level understanding of biology (36). As an example, a database dedicated to microarray data for microbial organisms—GenoScript (http://genoscript.pasteur.fr)—has recently been built using conceptual and technical basis analogous to those at work in GenoList, and thus constitutes an ideal target to undertake interoperability developments.
ACKNOWLEDGEMENTS
This work was supported by grants from the Réseau National des Génopoles. L.H. was partially funded by a contract with Danone Vitapole. We thank L.M. Jones for his help in optimizing the use of Sybase and in deploying the WebObjects application. We acknowledge support from S.T. Cole and input from A. Danchin and P. Glaser during the conception stage of the environment. We thank P. Casel, S. Grandino, H. Madaoui and J. Torrent for their contribution to the development of the GenoList application, and we are grateful to C. d’Enfert and T. Rossignol for fruitful discussions while setting up the CandidaDB database. Funding to pay the Open Access publication charges for this article was provided by Institut Pasteur.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb J-F, Dougherty BA, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Field D, Wilson G, van der Gast C. How do we compare hundreds of bacterial genomes? Curr. Opin. Microbiol. 2006;9:499–504. doi: 10.1016/j.mib.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Médigue C, Moszer I. Annotation, comparison and databases for hundreds of bacterial genomes. Res. Microbiol. 2007 doi: 10.1016/j.resmic.2007.09.009. doi:10.1016/j.resmic.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Field D, Feil EJ, Wilson GA. Databases and software for the comparison of prokaryotic genomes. Microbiology. 2005;151:2125–2132. doi: 10.1099/mic.0.28006-0. [DOI] [PubMed] [Google Scholar]
- 5.Gattiker A, Michoud K, Rivoire C, Auchincloss AH, Coudert E, Lima T, Kersey P, Pagni M, Sigrist CJ, et al. Automated annotation of microbial proteomes in SWISS-PROT. Comput. Biol. Chem. 2003;27:49–58. doi: 10.1016/s1476-9271(02)00094-4. [DOI] [PubMed] [Google Scholar]
- 6.Kersey P, Bower L, Morris L, Horne A, Petryszak R, Kanz C, Kanapin A, Das U, Michoud K, et al. Integr8 and Genome Reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alm EJ, Huang KH, Price MN, Koche RP, Keller K, Dubchak IL, Arkin AP. The MicrobesOnline Web site for comparative genomics. Genome Res. 2005;15:1015–1022. doi: 10.1101/gr.3844805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhuri RR, Pallen MJ. xBASE, a collection of online databases for bacterial comparative genomics. Nucleic Acids Res. 2006;34:D335–D337. doi: 10.1093/nar/gkj140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, Padki A, Zhao X, Dubchak I, Hugenholtz P, et al. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 2006;34:D344–D348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. The Comprehensive Microbial Resource. Nucleic Acids Res. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uchiyama I. MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups. Nucleic Acids Res. 2007;35:D343–D346. doi: 10.1093/nar/gkl978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 14.Moszer I. The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett. 1998;430:28–36. doi: 10.1016/s0014-5793(98)00620-6. [DOI] [PubMed] [Google Scholar]
- 15.Moszer I, Jones LM, Moreira S, Fabry C, Danchin A. SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res. 2002;30:62–65. doi: 10.1093/nar/30.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Médigue C, Viari A, Hénaut A, Danchin A. Colibri: a functional data base for the Escherichia coli genome. Microbiol. Rev. 1993;57:623–654. doi: 10.1128/mr.57.3.623-654.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 18.Tomb J-F, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 19.Boneca IG, de Reuse H, Epinat J-C, Pupin M, Labigne A, Moszer I. A revised annotation and comparative analysis of Helicobacter pylori genomes. Nucleic Acids Res. 2003;31:1704–1714. doi: 10.1093/nar/gkg250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang G, Ho C, Qiu Y, Cubas V, Yu Z, Cabau C, Cheung F, Moszer I, Danchin A. Specialized microbial databases for inductive exploration of microbial genome sequences. BMC Genomics. 2005;6:14. doi: 10.1186/1471-2164-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camus J-C, Pryor MJ, Médigue C, Cole ST. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 23.Ouzounis CA, Karp PD. The past, present and future of genome-wide re-annotation. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-comment2001. comment 2001.1–2001.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chetouani F, Glaser P, Kunst F. DiffTool: building, visualizing and querying protein clusters. Bioinformatics. 2002;18:1143–1144. doi: 10.1093/bioinformatics/18.8.1143. [DOI] [PubMed] [Google Scholar]
- 25.Perrière G, Duret L, Gouy M. HOBACGEN: database system for comparative genomics in bacteria. Genome Res. 2000;10:379–385. doi: 10.1101/gr.10.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chetouani F, Glaser P, Kunst F. FindTarget: software for subtractive genome analysis. Microbiology. 2001;147:2643–2649. doi: 10.1099/00221287-147-10-2643. [DOI] [PubMed] [Google Scholar]
- 29.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.d’Enfert C, Goyard S, Rodriguez-Arnaveilhe S, Frangeul L, Jones L, Tekaia F, Bader O, Albrecht A, Castillo L, et al. CandidaDB: a genome database for Candida albicans pathogenomics. Nucleic Acids Res. 2005;33:D353–D357. doi: 10.1093/nar/gki124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossignol T, Lechat P, Cuomo C, Zeng Q, Moszer I, d’Enfert C. CandidaDB: a multi-genome database for Candida species and related Saccharomycotina. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkm1010. (article to be published in the 2008 Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyer F, Morgat A, Labarre L, Pothier J, Viari A. Syntons, metabolons and interactons: an exact graph-theoretical approach for exploring neighbourhood between genomic and functional data. Bioinformatics. 2005;21:4209–4215. doi: 10.1093/bioinformatics/bti711. [DOI] [PubMed] [Google Scholar]
- 35.Stein LD. Integrating biological databases. Nat. Rev. Genet. 2003;4:337–345. doi: 10.1038/nrg1065. [DOI] [PubMed] [Google Scholar]
- 36.Ng A, Bursteinas B, Gao Q, Mollison E, Zvelebil M. Resources for integrative systems biology: from data through databases to networks and dynamic system models. Brief. Bioinform. 2006;7:318–330. doi: 10.1093/bib/bbl036. [DOI] [PubMed] [Google Scholar]