Abstract
The Worldwide Protein Data Bank (wwPDB; wwpdb.org) is the international collaboration that manages the deposition, processing and distribution of the PDB archive. The online PDB archive at ftp://ftp.wwpdb.org is the repository for the coordinates and related information for more than 47 000 structures, including proteins, nucleic acids and large macromolecular complexes that have been determined using X-ray crystallography, NMR and electron microscopy techniques. The members of the wwPDB–RCSB PDB (USA), MSD-EBI (Europe), PDBj (Japan) and BMRB (USA)–have remediated this archive to address inconsistencies that have been introduced over the years. The scope and methods used in this project are presented.
INTRODUCTION
The Worldwide Protein Data Bank (wwPDB) consists of organizations that act as deposition, data processing and distribution centers for PDB data. The members are the Research Collaboratory for Structural Bioinformatics (RCSB PDB), Macromolecular Structure Data Bank at the European Bioinformatics Institute (MSD-EBI), Protein Data Bank Japan (PDBj) and the BioMagResBank (BMRB) (1). Since 1971, the PDB has been responsible for the collection, processing, archiving and distribution of biological macromolecular structural data (2). Over the last 36 years, the archive has grown from seven structures to now more than 47 000. During this same period, the methods used to determine structures, the size of individual structures and the rate at which they are being solved have all changed, as have the ways in which the archive is used.
The methods used to collect, curate and process the data also have evolved over time. Different tools have been used to collect the data including the earliest version of AutoDep developed at Brookhaven (3), a reengineered version developed at MSD-EBI (4) and ADIT used by RCSB PDB and PDBj (5,6). Over the years, data curation has become more and more automated, although expert curators still review the structures to ensure they are represented correctly. Finally, there have been subtle but definite changes in the PDB file format (7) and the definitions for the various records have been subject to different interpretations both by depositors and by curators. The result of all of these factors has been inconsistencies and outright errors in the data.
The wwPDB therefore undertook a project to remediate the entire archive. The scope of this remediation project has been to address problems that limit the utility of the archive as a whole. Thus, we have focused on the following areas: (i) improving the detailed chemical description and nomenclature of the monomer units of the biological polymers and small molecule ligands; (ii) resolving any remaining differences between the chemical and the macromolecular sequences, and updating sequence database references and taxonomies; (iii) improving the representation of viruses; and (iv) verifying primary citation assignments. We also addressed miscellaneous errors, some REMARKS, and structure factor and NMR restraint data. Coordinates have not been changed.
The impact of this work on the data files and dictionaries produced by the wwPDB are described in the following sections.
CHEMICAL DESCRIPTIONS: THE CHEMICAL COMPONENT DICTIONARY
A major portion of the wwPDB remediation project has been devoted to improving the chemical description and nomenclature used in the annotation of macromolecular structure data. This work has been incorporated into a new reference dictionary called the Chemical Component Dictionary. Key features include:
Model and idealized coordinates
Chemical descriptors (e.g. SMILES (8) and InChI (9)) and systematic names
Stereochemical assignments and aromatic bond assignments
IUPAC nomenclature for standard amino acids and nucleotides (10) with the exception of the well-established convention for C-terminal atoms OXT and HXT
More conventional atom labeling
Removal of redundant ligands
Additional description of protonation states
The remediated dictionary of chemical components provides a richer and more accurate description of each molecule. The more detailed chemical definitions have been used to recheck the assignments of the monomer (13M+) and non-polymer (170K+) molecules in the PDB archive. While this chemical reference dictionary has been used in the remediation of each PDB entry, much of the information in this dictionary is not directly incorporated within individual remediated entries. In particular, the expressivity of the chemical description within PDB format CONECT records is very limited. PDB users are encouraged to take direct advantage of the content of the new chemical dictionary.
Additional chemical definitions have been created for amino acids in different states of protonation. These definitions document the nomenclature for the additional protons not specified in the standard definitions. The additional definitions are maintained in a Companion Amino Acids Variants Dictionary that provides complete molecular definitions of the protonated amino acids (Table 1).
Table 1.
Histidine Variants in the Companion Amino Acids Variants Dictionary
| CODE | Variant |
|---|---|
| HIS | HISTIDINE |
| HIS_LEO2 | L-HISTIDINE C-TERMINAL DEPROTONATED FRAGMENT |
| HIS_LEO2H | L-HISTIDINE C-TERMINAL PROTONATED FRAGMENT |
| HIS_LEO2H_DHD1 | L-HISTIDINE-C-TERMINAL PROTONATED FRAGMENT/WITH SIDE CHAIN DEPROTONATED ND1 |
| HIS_LEO2H_DHE2 | L-HISTIDINE-C-TERMINAL PROTONATED FRAGMENT/WITH SIDE CHAIN DEPROTONATED NE2 |
| HIS_LEO2_DHD1 | L-HISTIDINE-C-TERMINAL DEPROTONATED FRAGMENT/WITH SIDE CHAIN DEPROTONATED ND1 |
| HIS_LEO2_DHE2 | L-HISTIDINE-C-TERMINAL DEPROTONATED FRAGMENT/WITH SIDE CHAIN DEPROTONATED NE2 |
| HIS_LFOH | L-HISTIDINE FREE NEUTRAL |
| HIS_LFOH_DHD1 | L-HISTIDINE-FREE NEUTRAL/WITH SIDE CHAIN DEPROTONATED ND1 |
| HIS_LFOH_DHE2 | L-HISTIDINE-FREE NEUTRAL/WITH SIDE CHAIN DEPROTONATED NE2 |
| HIS_LFZW | L-HISTIDINE FREE ZWITTERION |
| HIS_LFZW_DHD1 | L-HISTIDINE-FREE ZWITTERION/WITH SIDE CHAIN DEPROTONATED ND1 |
| HIS_LFZW_DHE2 | L-HISTIDINE-FREE ZWITTERION/WITH SIDE CHAIN DEPROTONATED NE2 |
| HIS_LL | L-HISTIDINE - LINKING EMBEDDED FRAGMENT |
| HIS_LL_DHD1 | L-HISTIDINE-LINKING EMBEDDED FRAGMENT/WITH SIDE CHAIN DEPROTONATED ND1 |
| HIS_LL_DHE2 | L-HISTIDINE-LINKING EMBEDDED FRAGMENT/WITH SIDE CHAIN DEPROTONATED NE2 |
| HIS_LSN3 | L-HISTIDINE N-TERMINAL PROTONATED FRAGMENT |
| HIS_LSN3_DHD1 | L-HISTIDINE-N-TERMINAL PROTONATED FRAGMENT/WITH SIDE CHAIN DEPROTONATED ND1 |
| HIS_LSN3_DHE2 | L-HISTIDINE-N-TERMINAL PROTONATED FRAGMENT/WITH SIDE CHAIN DEPROTONATED NE2 |
Note: The Chemical Component Dictionary is accompanied by a companion dictionary of amino acid variants that provides additional nomenclature information for the protonation states of standard amino acids in N-terminal, C-terminal and free forms. This dictionary also includes common side chain protonation states. It is similar to residue variants used in modeling software such as Charmm (42) and Amber (43).
The Chemical Component Dictionary provided the basis for the remediation of all monomer units and small molecule ligands in the PDB files. The impact of the new chemical definitions is seen in the atoms names, atom types, residue names and residue assignments.
The dictionaries and detailed descriptions of the improved description of chemical components are available for download from http://www.wwpdb.org.
CHANGES TO THE PDB COORDINATE ENTRIES
Atom and residue naming
Atom names in the polymer chains (ATOM records in the PDB file format) in the remediated data files directly reflect the nomenclature changes in the chemical dictionary. These names uniformly begin with their atom type symbol, including hydrogen atoms. Names beginning with numbers and unusual atom names have been changed accordingly. Atom types are provided for every atom (i.e. ATOM record columns 77–78), so prior atom name justification conventions should no longer be assumed in reading atom names. As with the Chemical Component Dictionary, names for standard amino acids and nucleotides follow IUPAC recommendations (10) with the exception of the well-established convention for C-terminal atoms OXT and HXT. These nomenclature changes have been applied to standard polymeric chemical components only.
In the remediated entries, the atom names in the Companion Amino Acids Variants Dictionary have been used to describe protonated molecules; however, the extended residue names are not used. The proton names are assigned to the standard residue (i.e. HIS).
Residue assignments have all been rechecked against the new and more detailed chemical reference dictionary. A residue assignment was changed in the remediated entry if it was inconsistent with chemical connectivity and/or stereochemistry of its prior assignment, or the prior assignment was obsoleted.
DNA and RNA nucleotides now have separate chemical definitions. The DNA and RNA nucleotides are distinguished with the DNA forms relabeled as DA, DC, DG and DT. The nucleotide atom nomenclature has been standardized, and the format of the ATOM record provides explicit atom type information. Modified nucleotides formerly identified as using the ‘plus-nucleotide’ syntax have been relabeled with the particular 3-letter code corresponding to the full-modified nucleotide definition (Table 2).
Table 2.
RNA and DNA atom names in the remediated and unremediated files
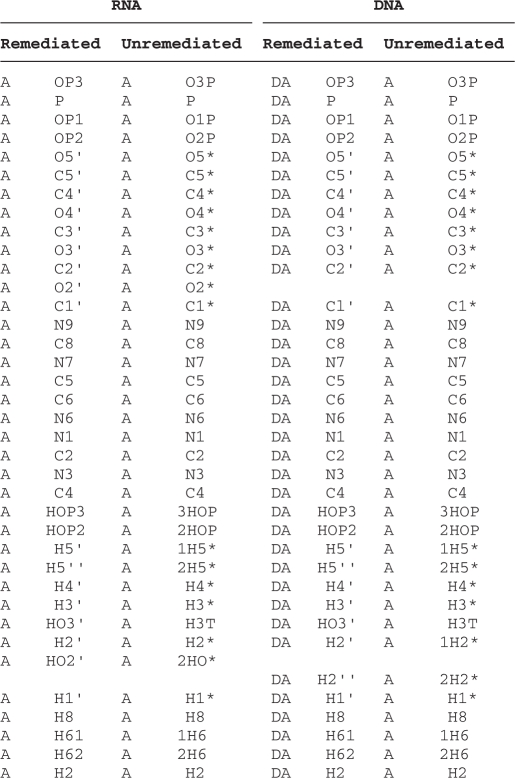 |
The remediated and unremediated residue name and atom name are given for the linked adenosine residue in RNA and DNA.
The impact of the changes in the Chemical Component Dictionary on ligands (HET groups) in PDB entries consisted of removing redundant definitions, absorbing small modifying functional groups into complete components, and removing definitions with ambiguous chemical descriptions. More than 170 000 ligands in the data files were checked against the dictionary, and as a result 7700 names changed and 330 component definitions were obsoleted. The obsolete chemical components remain in the dictionary with an identifying status of ‘OBS’. Beyond ensuring that atom names begin with their type symbol, no attempt was made to extend systematic nomenclature to non-polymer chemical components.
Examples of obsolete heterogroup names
The various hydrated magnesium ions (MO1, MO2, MO3, MO4, MO5 and MO6) have been split into an MG (magnesium ion) and the appropriate number of water molecules. In a similar manner, other examples are now obsolete het-groups. KO4 has been split into a potassium ion (K) and four water molecules (HOH), while het-group 543 has been split into a CA (calcium ion), an EOH (ethanol molecule) and six water molecules. Some 64 such groups were made obsolete. Other groups have been superceded to give a single unique het-group name in the PDB collection, including: LTR, now TRP (L-Tryptophan); FCY, now CYS (cysteine); NEV and NIV, replaced by NVP (Nevirapine); and GS4, replaced by SGC (4-thio-beta-D-glucopyranose). More than 180 such groups were made obsolete. Where possible, single atom or small groups have been replaced by complex single compound entries. These include making the ethyl group (ETH) obsolete and creating new hetgroups where previous PDB entries contained an ETH linked to another het-group.
Sequence and taxonomy
Some inconsistencies between the chemical and the coordinate macromolecular sequences were largely resolved in data files deposited before 1998 when the first set of mmCIF data files were released in 2000 (11). Remaining differences between the chemical and the macromolecular sequence have been resolved through the remediation project. All of these changes have been applied to the remediated files in PDB format. The remediated data files deposited pre-1998 reflect many changes in SEQRES and ATOM records that were required to resolve inconsistencies. Typical changes included: assignment of poly-ALA sequences to the corresponding amino acids in the chemical sequence, reassignment of chain identifiers to correspond to complete chemical sequences, correcting terminal atom nomenclature at internal gaps and providing non-blank labels for all polymer chains.
Sequence database references and all associated difference records have been checked and/or updated along with associated taxonomy information for ∼61 K sequences. UniProt (12) references have been used where possible. Sequence database correspondences were verified in December 2006. To maintain these correspondences in the future, the PDB will use the mapping data from the Structure Integration with Function, Taxonomy and Sequence (SIFTS) initiative (13).
Virus representation
The representation of viruses and large assemblies has been extended to better describe existing and anticipated entries of this type. The description of the deposited and experimental coordinate frames, symmetry and frame transformations has been generalized to better represent experiments that do not exclusively use crystallographic symmetry. This description has been properly decoupled from the description of non-crystallographic symmetry (NCS) exploited within a crystallographic structure determination. A simplified notation has been adopted to express the symmetry generation of assemblies from deposited coordinates and a standard set of matrix operations describing either point, helical or crystallographic symmetry.
Errors in archived transformation matrices required to build full assemblies from the deposited coordinates for the existing 280+ virus structures were identified by inspection of images generated with the multiscale model module of UCSF Chimera (14). Corrected matrices were obtained from the Virus Particle Explorer database (VIPERdb, http://viperdb.scripps.edu) (15) or the Protein Quaternary Structure server (PQS, http://pqs.ebi.ac.uk) (16). The corrected transformation matrices are included in remediated PDB format files.
In addition, transformations to crystal frame were collected from author text remarks or primary citations, or they were extracted from SCALE records for ∼210 icosahedral virus crystal structures. NCS operations defining crystal asymmetric units were determined and crystal packing was inspected using the crystal contacts module of UCSF Chimera. Entries with structure factors were validated with SFCHECK (17). For structures deposited in the crystal frame, NCS operations are provided in the MTRIX records; for structures deposited in other frames, a text description of how to build the crystal asymmetric unit is provided in REMARK 285.
Primary citations
All primary citations have been rechecked. Citations formerly marked as To Be Published have been researched and either the citation has been identified or marked as Not Published. PubMed identifiers have been provided where available. The PubMed identifiers only appear in the remediated mmCIF and PDBML files.
Miscellaneous improvements in consistency
To improve the overall consistency and accuracy of the archive, a variety of individual corrections have been applied. These include beamline names, synchrotron facility names, source organism, method names, elimination of singleton alternate atom location labels, diffraction wavelength, computing methods and the correction of miscellaneous typographical errors. The latter includes correcting misspellings and nonstandard usage, resolving of duplicated identifiers (e.g. author residue numbers, entity and citation identifiers) and properly distinguishing null values from zero.
Free text PDB REMARKS have generally not been remediated and have not been incorporated in the remediated PDB entries. These remarks remain in a legacy remark category data_PDB_remark in the mmCIF and PDBML remediated files. These remarks can also be viewed in the original entries that will always be preserved.
The following PDB remarks are constructed from text templates using data items in the mmCIF/PDBML entry file: 2, 3, 4, 100, 200, 210, 215, 220, 225, 230, 240, 245, 247, 250, 265, 280, 290, 300, 350, 375, 465, 470, 500, 525, 900. These PDB remarks are reports constructed from the individual data items in the more structured mmCIF/PDBML data files. While the information presented in the PDB remarks directly corresponds to the content of the mmCIF/PDBML data files, the content of the PDB remark may not be comprehensive. The mmCIF/PDBML files should be used to obtain the most complete view of a data entry. For instance, X-ray data collection details in REMARK 200 may be found in the mmCIF/PDBML data categories in refln_group category group, and X-ray refinement details in REMARK 3 may be found in the data categories in the refine_group category group.
STRUCTURE FACTOR AND NMR RESTRAINT DATA
Many issues with structure factor data files have been addressed through a collaboration with the developers of the Uppsala Electron Density Server (EDS) (18).
Nomenclature standardization for NMR restraint files in the current PDB archive has been done as part of the NMR Restraints Grid Project, a collaboration with the Collaborative Computing Project for NMR and Bijvoet Center for Biomolecular Research. NMR restraint data files with atom nomenclature corresponding to remediated PDB data files will be available by the end of 2007.
FORMATS
The focus of the remediation project has been to address certain data consistency issues within entries and to bring all of the files in the archive to the current level of each of the PDB data formats (PDB, mmCIF/PDBx and PDBML). While the content of certain records may reflect changes from remediation, the syntax and organization of this information is largely the same as for new entries processed by PDB. Some changes in content may affect the way in which existing records are used; these issues for particular formats are discussed below.
PDB format
The record structure of the PDB format is essentially unchanged by the remediation project. The format prior to the remediation project was documented in the PDB V2.3 contents guide (19). The small number of format differences for the remediated entries are documented in the PDB V3.0.1 contents guide (http://www.wwpdb.org/docs.html). There are a few issues related to the use of the remediated files that may require attention of software developers. These include:
Standardization of hydrogen atom nomenclature has required clarifying historical conventions in the justification of atom names in PDB ATOM records. These conventions were used to convey atom type information in early PDB format entries in which the element symbol was not included. The remediated entries uniformly include atom type information in columns 77–78. Using the justification of the atom name to derive atom type information is now strongly discouraged.
DNA nucleotide residues are differentiated from RNA nucleotides in the remediated data files. DNA residues are now preceded by the letter ‘D’ (e.g. DA, DC and DG). Nucleotide modifications in the remediated files are now fully described as complete chemical components. The prior practice of identifying a nucleotide modification of with a preceding ‘plus’ character is not used.
To distinguish PDB files containing the remediated nomenclature from previous files, REMARK 4 has been updated to reflect the format version 3.0 and a notation that the file has been remediated.
mmCIF/PDBx format
The remediated data files introduce no change in the syntax of mmCIF format data files (20). The following issues may require the attention of software developers:
The maximum line length used in writing the remediated data files has been extended such that each atom record in the atom_site category is written in a single line.
Additional auditing information is included in each remediated file. The underlying dictionary name, location and version are included in category audit_conform. Version information for each mmCIF data file is included in category pdbx_version.
The definitions of the data items included in the remediated mmCIF files is described in the PDB exchange dictionary version 1.045 (PDBx) (21) (http://mmcif.pdb.org/dictionaries/ascii/mmcif_pdbx.dic) This version of the dictionary incorporates some improvements in the consistency of data typing, corrections to category key structure, and miscellaneous corrections in definitions, examples and enumerations. The details of the changes are described in the dictionary history.
PDBML-XML
The remediated PDBML-XML (22) files are translated from mmCIF remediated data files and reflect the content changes described in the previous section. The revised PDBML XSD schema also includes all the changes in the PDBx version 1.045. The changes in category key structure (e.g. citation_author, refine_ls_restr_ncs) and some data type changes may require attention in parsing software.
METHODS
Chemical dictionary
The approach to improving the chemical description in the PDB relied heavily on improving and verifying the Chemical Component Dictionary. A chemical component description consists of a representative 3-dimensional model taken from the archive along its associated atom nomenclature, covalent bonding and stereochemistry.
This work involved extracting all instances of each chemical component from the archive and verifying the chemical assignments. Since prior chemical definitions did not include detailed stereochemical assignments, these were first verified relative to the molecular name. New stereochemically specific chemical definitions where created in cases where multiple enantomeric forms had previously been assigned to the same component identifier.
The preliminary screening of chemical component definitions took advantage of the stereochemical assignments used by MSDCHEM (23) obtained using the CACTVS (24) chemical informatics toolset. The stereochemical and aromatic bond assignments for the complete chemical dictionary were later rechecked using CACTVS tools and the OpenEye OEChem tools (25). Software assisted assignments of stereochemistry and aromaticity are limited to the chemical systems for which these tools were developed (e.g. primarily tetravalent organic systems). Saccharide components were also checked using the GlycoSciences PDB-care software tool (26). Improved description of chemical components involving metal coordination or dative bonding is ongoing.
A set of computationally modeled coordinates was provided in each component definition if a satisfactory set of coordinates could be obtained using either CORINA (27) or OpenEye Omega (28) packages. Systematic chemical names and chemical descriptors were also included in each component definition. Systematic names were computed using ACDLabs ACD/Name batch naming software (29) and OpenEye Lexichem (25). Stereo SMILES descriptors were computed using both CACTVS and OpenEye tools, and InChI descriptors were computed using software distributed by this IUPAC project.
The improved chemical component definitions were then used to recheck the assignments of each non-polymer, modified amino acid or modified nucleotide component instance in the PDB archive. This work involved extracting the coordinates of each component, deriving the chemical connectivity of the component, and comparing this to the chemical dictionary. This process was driven by the DOHLC data processing program that uses BALI (30) and OpenBabel (31,32) for bond assignment and subgraph matching software from the CCP4 Coordinate Library (23).
Integration of remediated data
To manage and track data files during the remediation project a CVS archive was created for released PDB entries as of March 2006. The CVS repository was built from the mmCIF versions of these released entries.
Because the changes in sequence and taxonomy manifest the greatest change in the organization of an entry, these remediation corrections were integrated first. This work and other integration operations were performed using tools adapted from the RCSB PDB's data processing and annotation software suite (5,6,33). These tools perform edits in the macromolecular sequence and propagate these changes consistently throughout the entry. Sequence database correspondences and updated taxonomy information were also updated at this point.
After revisions in macromolecular sequence were applied, changes in component-level (modified residue and ligand) nomenclature were reintegrated into the remediated entries. Primary citation data, revised virus representations, corrections to experimental and other data items were then integrated.
Atom-level nomenclature changes were performed in the final software translation step prior to creating remediated files in PDB and PDBML formats. This was done in order to allow atom nomenclature to be refined during the course of the project. Beginning in December 2006, remediated data files in PDB, mmCIF/PDBx and PDBML formats along with supporting dictionaries were provided for public review.
Testing and validation
After all of the content and corrections were integrated into the remediated data files, these files were rechecked for consistency. Each of the wwPDB partners has contributed to this final validation of the remediated data files by applying their respective data processing and database tools to this task.
Using PDBx as a reference, each of the remediated mmCIF files was rechecked. This dictionary-level testing identifies inconsistencies in controlled vocabularies, boundary conditions and relationships between common identifiers. Similar checks of this type were performed on the PDBML data files using the XML schema translated from the PDBx. Checks for atom and residue nomenclature consistency were also performed against the Chemical Component Dictionary.
Data files were loaded into several relational database systems with different table schema. These loading operations provided further tests of data type, controlled vocabulary, boundary value and referential integrity. Loading data within a native XML database system provided additional complementary diagnostics.
During the public review of the remediated data, we benefited greatly from diagnostics contributed from PDB users who exercised the remediated data files in the application area of visualization, crystallographic phasing and refinement, docking, and homology modeling. Questions and comments about the remediated data should be sent to info@wwpdb.org.
Software support
In producing the remediated PDB data files, every effort was made to minimize the impact of the remediation on existing software applications. However, in order to support community standard nomenclature, Version 3.0 of the PDB Format was introduced. While adopting more standard nomenclature greatly simplifies the use and comparison of PDB data in most respects, many existing software applications have been developed to cope with the eccentric historical nomenclature.
As described in the previous section on ‘Testing and Validation’, the remediation project has included active participation from PDB users and software developers. The wwPDB maintained an informational website and mail server during the last year of the project to provide project information to earlier adopters and testers. The wwPDB also hosted a workshop for software developers at the 2007 American Crystallographic Association's annual meeting to address data representation issues that became highlighted during the remediation project.
By the time the remediated data files replaced the existing entries in August 2007, many widely-used visualization programs such as OpenRasMol, Chimera, PyMol, JMol, WebMol, KiNG, the Molecular Biology Toolkit, jV (formerly known as PDBjViewer) and Discovery Studio Visualizer were already compatible with the remediated PDB data format (34–41). wwPDB and user-contributed tools are also available to translate between the nomenclatures used in old and remediated data formats. A current list of applications reported as compatible with the remediated data files and related conversion software tools is available at http://remediation.wwpdb.org/software.html. All of the wwPDB deposition sites continue to accept depositions with either nomenclature.
FTP
The remediated data and data annotated and released by members of the wwPDB are available for download from ftp://ftp.wwpdb.org. This site is updated on a weekly basis.
A snapshot of the unremediated PDB archive (as of July 31, 2007) is available at ftp://ftp.rcsb.org. This site has been frozen, and will not be updated.
ACKNOWLEDGEMENTS
The contributions of all of the wwPDB staff members are gratefully acknowledged. Special thanks goes to the many PDB users who tested the remediated data and provided comments, especially Dan Bolser, Alexandre M.J.J. Bonvin, Tommy Carstensen, Roland Dunbrack, Howard Feldman, Dave Howorth, Miron Livny, Eric Pettersen, the Richardson Lab at Duke University and Clemens Vonrhein. The RCSB PDB is operated by Rutgers, The State University of New Jersey and the University of California, San Diego. It is supported by funds from the National Science Foundation, the National Institute of General Medical Sciences, the Office of Science, Department of Energy, the National Library of Medicine, the National Cancer Institute, the National Center for Research Resources, the National Institute of Biomedical Imaging and Bioengineering, National Institute of Neurological Disorders and Stroke and the National Institute of Diabetes and Digestive and Kidney Diseases. The EMBL-EBI MSD group gratefully acknowledges the support of the Wellcome Trust, the EU (FELICS, EXTENDNMR, EuroCarbDB and 3DEM), the BBSRC, the MRC and EMBL. PDBj is supported by grant-in-aid from the Institute for Bioinformatics Research and Development, Japan Science and Technology Agency (BIRD-JST), and the Ministry of Education, Culture, Sports, Science and Technology (MEXT). The BMRB is supported by NIH grant LM05799 from the National Library of Medicine. Funding to pay the Open Access publication charge was provided by NSF DBI 03-12718.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berman HM, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein FC, Koetzle TF, Williams GJB, Meyer E.F., Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 3.Lin D, Manning NO, Jiang J, Abola EE, Stampf D, Prilusky J, Sussman JL. AutoDep: a web-based system for deposition and validation of macromolecular structural information. Acta Cryst. D. 2000;D56:828–841. doi: 10.1107/s0907444900005655. [DOI] [PubMed] [Google Scholar]
- 4.Keller PA, Henrick K, McNeil P, Moodie S, Barton GJ. Deposition of macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 1998:1105–1108. doi: 10.1107/s0907444998008464. [DOI] [PubMed] [Google Scholar]
- 5.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutta S, Burkhardt K, Bluhm WF, Berman HM. Using the tools and resources of the RCSB Protein Data Bank. Current Protocols in Bioinformatics. 2005:1.9.1–1.9.40. doi: 10.1002/0471250953.bi0109s20. [DOI] [PubMed] [Google Scholar]
- 7.Westbrook J, Fitzgerald PM. The PDB format, mmCIF formats and other data formats. In: Bourne PE, Weissig H, editors. Structural Bioinformatics. NJ, Hoboken: John Wiley & Sons, Inc.; 2003. pp. 161–179. [Google Scholar]
- 8.Weininger D. SMILES 1. Introduction and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31. [Google Scholar]
- 9.© The International Union of Pure and Applied Chemistry. IUPAC International Chemical Identifier (InChI) 2005. (contact: secretariat@iupac.org)
- 10.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the standardization of data bases of protein and nucleic acid structures determined by NMR spectroscopy. J. Biomol. NMR. 1998;12:1–23. doi: 10.1023/a:1008290618449. [DOI] [PubMed] [Google Scholar]
- 11.Bhat TN, Bourne P, Feng Z, Gilliland G, Jain S, Ravichandran V, Schneider B, Schneider K, Thanki N, Weissig H, et al. The PDB data uniformity project. Nucleic Acids Res. 2001;29:214–218. doi: 10.1093/nar/29.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velankar S, McNeil P, Mittard-Runte V, Suarez A, Barrell D, Apweiler R, Henrick K. E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res. 2005;33:D262–D265. doi: 10.1093/nar/gki058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novoselov KP, Shirabaikin DB, Umanskii SY, Vladimirov AS, Minushev A, Korkin AA. CHIMERA: a software tool for reaction rate calculations and kinetics and thermodynamics analysis. J. Comput. Chem. 2002;23:1375–1389. doi: 10.1002/jcc.10105. [DOI] [PubMed] [Google Scholar]
- 15.Shepherd CM, Borelli IA, Lander G, Natarajan P, Siddavanahalli V, Bajaj C, Johnson JE, Brooks C.L., III, Reddy VS. VIPERdb: a relational database for structural virology. Nucleic Acids Res. 2006;34:D386–D389. doi: 10.1093/nar/gkj032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henrick K, Thornton JM. PQS: a protein quarternary file server. Trends Biochem. Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 17.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 18.Kleywegt GJ, Harris MR, Zou JY, Taylor TC, Wahlby A, Jones TA. The Uppsala electron-density server. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2240–2249. doi: 10.1107/S0907444904013253. [DOI] [PubMed] [Google Scholar]
- 19.Callaway J, Cummings M, Deroski B, Esposito P, Forman A, Langdon P, Libeson M, McCarthy J, Sikora J, et al. Brookhaven National Laboratory; 1996. Protein data bank contents guide: atomic coordinate entry format description. [Google Scholar]
- 20.Fitzgerald PMD, Westbrook JD, Bourne PE, McMahon B, Watenpaugh KD, Berman HM. Definition and exchange of crystallographic data. In: Hall SR, McMahon B, editors. International Tables for Crystallography. G. Dordrecht, The Netherlands: Springer; 2005. pp. 295–443. [Google Scholar]
- 21.Westbrook J, Henrick K, Ulrich EL, Berman HM. Definition and exchange of crystallographic data. In: Hall SR, McMahon B, editors. International Tables for Crystallography. G. Dordrecht, The Netherlands: Springer; 2005. pp. 195–198. [Google Scholar]
- 22.Westbrook J, Ito N, Nakamura H, Henrick K, Berman HM. PDBML: the representation of archival macromolecular structure data in XML. Bioinformatics. 2005;21:988–992. doi: 10.1093/bioinformatics/bti082. [DOI] [PubMed] [Google Scholar]
- 23.Golovin A, Oldfield TJ, Tate JG, Velankar S, Barton GJ, Boutselakis H, Dimitropoulos D, Fillon J, Hussain A, Ionides JM, et al. E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res. 2004;32(Database issue):D211–D216. doi: 10.1093/nar/gkh078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ihlenfeldt W, Takahasi Y, Abe H, Sasaki S. Computation and management of chemical properties in CACTVS: An extensible networked approach toward modularity and flexibility. J. Chem. Inf. Comp. Sci. 1994;34:109–116. [Google Scholar]
- 25.OpenEye Scientific Software Inc. OpenEye OEChem version 1.5. 2007. www.eyesopen.com Santa Fe, NM, USA.
- 26.Lutteke T, von der Lieth CW. pdb-care (PDB carbohydrate residue check): a program to support annotation of complex carbohydrate structures in PDB files. BMC Bioinformatics. 2004;5:69. doi: 10.1186/1471-2105-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasteiger J, Rudolph C, Sadowski J. Automatic generation of 3D-atomic coordinates for organic molecules. Tetrahedron Comp. Method. 1990;3:537–547. [Google Scholar]
- 28.OpenEye Scientific Software Inc. OpenEye Omega version 2.2.1. 2007. www.eyesopen.com Santa Fe, NM, USA.
- 29.Advanced Chemistry Development, I. ACD/Name Batch, version 9.0, Toronto ON, Canada. 2007. www.acdlabs.com.
- 30.Hendlich M, Rippmann F, Barnickel G. BALI: automatic assignment of bond and atom types for protein ligands in the Brookhaven Protein Databank. J. Chem. Inf. Comp. Sci. 1997;37:774–778. [Google Scholar]
- 31.Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, Wegner J, Willighagen EL. The blue obelisk-interoperability in chemical informatics. J. Chem. Inf. Model. 2006;46:991–998. doi: 10.1021/ci050400b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Open Babel Package. Version 2.0.1. 2006. http://openbabel.sourceforge.net/
- 33.Westbrook J, Feng Z, Burkhardt K, Berman HM. Validation of protein structures for the Protein Data Bank. Meth. Enz. 2003;374:370–385. doi: 10.1016/S0076-6879(03)74017-8. [DOI] [PubMed] [Google Scholar]
- 34.Sayle R, Milner-White EJ. RasMol: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 35.Bernstein HJ. Recent changes to RasMol, recombining the variants. Trends Biochem. Sci. 2000;25:453–455. doi: 10.1016/s0968-0004(00)01606-6. [DOI] [PubMed] [Google Scholar]
- 36.DeLano W. The PyMOL Molecular Graphics System on World Wide Web. 2002. http://www.pymol.org.
- 37.Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/
- 38.Walther D. WebMol–a Java-based PDB viewer. Trends Biochem. Sci. 1997;22:274–275. doi: 10.1016/s0968-0004(97)89047-0. [DOI] [PubMed] [Google Scholar]
- 39.Davis IW, Arendall W.B., III, Richardson DC, Richardson JS. The backrub motion: how protein backbone shrugs when a sidechain dances. Structure. 2006;14:265–274. doi: 10.1016/j.str.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 40.Moreland JL, Gramada A, Buzko OV, Zhang Q, Bourne PE. The molecular biology toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics. 2005;6:21. doi: 10.1186/1471-2105-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinoshita K, Nakamura H. eF-site and PDBjViewer: database and viewer for protein functional sites. Bioinformatics. 2004;20:1329–1330. doi: 10.1093/bioinformatics/bth073. [DOI] [PubMed] [Google Scholar]
- 42.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 43.Weiner P, Kollman P. Amber. J. Comput. Chem. 1981;2:287–303. [Google Scholar]


