Abstract
TreeFam (http://www.treefam.org) was developed to provide curated phylogenetic trees for all animal gene families, as well as orthologue and paralogue assignments. Release 4.0 of TreeFam contains curated trees for 1314 families and automatically generated trees for another 14 351 families. We have expanded TreeFam to include 25 fully sequenced animal genomes, as well as four genomes from plant and fungal outgroup species. We have also introduced more accurate approaches for automatically grouping genes into families, for building phylogenetic trees, and for inferring orthologues and paralogues. The user interface for viewing phylogenetic trees and family information has been improved. Furthermore, a new perl API lets users easily extract data from the TreeFam mysql database.
INTRODUCTION
Biologists studying a gene in one model organism often wish to transfer functional information between species. To do this, it is essential to know how the gene is related to other genes in a family. Using a phylogenetic tree, it is possible to infer orthologues—related genes in different species that diverged at the time of a speciation event—and paralogues, that is related genes that originated via a duplication event within a species (1).
In his original definition of orthology, Fitch defined orthologues in terms of a phylogenetic tree of a gene family (1). It has now been well established that analysis of phylogenetic trees is a very accurate way to determine orthology (2,3), which led us to develop the TreeFam database and accompanying website in 2005 (4). TreeFam aims to be a curated database of phylogenetic trees of all animal gene families, focusing on gene sets from animals with completely sequenced genomes. In TreeFam, orthologues and paralogues are inferred from the phylogenetic tree of a gene family. Tree-based inference of orthologues is more robust to rate differences than blast-based orthologue inference, which has been used in other databases such as InParanoid (5), KOGs (6), HomoloGene (7) and OrthoMCL-DB (8). Furthermore, tree-based results can be easily visualized and for some purpose are more informative, since gene losses and duplications can be inferred and dated on a tree.
In addition to the databases mentioned above, many other databases provide animal gene families on the genome-wide scale, such as PANTHER (9), Phylofacts (10), PhIGs (11) and SYSTERS (12). They usually display the phylogenetic trees, but most do not computationally infer orthologues from the gene trees. Like TreeFam, a few databases explicitly predict orthologues based on phylogenetic trees. These include HOGENOM (13) and PhylomeDB (14). While HOGENOM allows users to calculate the orthologues on the fly with a program that connects to their database, PhylomeDB presents orthologues as directly searchable results. Furthermore, Ensembl now collaborates with TreeFam, and uses the same tree-building and orthologue inference algorithms (15). It is clear that the tree-based methods are theoretically attractive, but building accurate gene trees remains a major challenge.
In this update, we have expanded TreeFam to include 25 fully sequenced animal genomes and four outgroup genomes. Furthermore, we have made many software improvements since the first release of TreeFam. These include (i) new algorithms for phylogenetic inference, (ii) a more user-friendly website and (iii) a perl interface (API) to the publicly available database. Together with the new features, TreeFam is an even more useful resource for identifying orthologues and paralogues in animal species and for studying evolution of animal gene families.
MATERIALS AND METHODS
Sequence data
Seventeen new species have been added since TreeFam v1 (4). TreeFam v4 contains predicted protein sequences from the fully sequenced genomes of 25 animal species: human, chimpanzee, macaque, mouse, rat, cow, dog, opossum, chicken, frog, two pufferfish (Takifugu and Tetraodon), zebrafish, medaka, stickleback, sea squirts (Ciona intestinalis and C. savignyi), two fruit-flies (Drosophila melanogaster and D. pseudoobscura), two mosquitoes (Aedes aegypti and Anopheles gambiae), the flatworm Schistosoma mansoni, and the nematodes Caenorhabditis elegans, C. briggsae and C. remanei. In addition, four outgroup genomes are included: baker's yeast, fission yeast, rice and thale cress (Arabidopsis).
The C. briggsae and C. remanei proteins were downloaded from WormBase (16), D. pseudoobscura proteins from FlyBase (17), fission yeast and flatworm proteins from GeneDB (18), thale cress proteins from TIGR (19), rice proteins from the Beijing Genomics Institute (20) and the remaining sequences from Ensembl (15). In addition to these species, TreeFam includes UniProt (21) proteins from animal species whose genomes have not been fully sequenced. For TreeFam v4, all sequences were downloaded in October 2006.
Overall strategy
TreeFam is a two-part database: a first part consisting of automatically generated trees (TreeFam-B) and a second part that consists of manually curated trees (TreeFam-A).
Automatically generating trees for TreeFam-B
TreeFam v1 used clusters of genes from PhIGs (11) as seeds for B families. However, for TreeFam v4, each B seed consists of genes from ‘core’ species from the corresponding TreeFam-3 family. ‘Core’ species are those selected to have high-quality reference genome sequences and gene predictions with good phylogenetic representation of the phyla of biological or phylogenetic importance. These were human, mouse, opossum, chicken, frog, pufferfish (Takifugu), zebrafish, sea squirt (C. savignyi), flatworm (22), D. melanogaster, C. elegans, baker's yeast, fission yeast, thale cress and rice. This change allowed TreeFam to use new gene sets that are absent from PhiGs, and to ensure that families remain stable from one release to the next.
Each seed family in TreeFam-B is expanded by using blast and hmmer to search for sequence matches among the animal and outgroup protein data sets, including animal sequences from UniProt. In TreeFam v1, we expanded each seed to form a full family. In TreeFam v4, we also made a ‘clean’ family from each seed, which only contains genes from fully sequenced genomes. The reasons for making a clean family were that (i) truncated proteins from UniProt sometimes cause problems for tree-building algorithms, and (ii) the algorithms we use to build trees (described subsequently) perform best when given both DNA and protein sequences, but many UniProt proteins lack easily identifiable DNA sequences.
Furthermore, for TreeFam v4, we employed a new approach to ensure that each animal gene only appears in one family. First, we assigned each transcript to the B or A family for which it had the highest-scoring hmmer match. Second, for each family, we only kept one transcript from each gene: the transcript with the highest-scoring hmmer match to the family. The one situation in which a gene is allowed to belong to more than one family is where the gene has transcripts with highest-scoring matches to different families. This can occur because EnsEMBL takes all the overlapping transcripts as one gene, whereas bad gene predictions or true gene fusion events may lead to transcripts that only share short fragments at the DNA level and have different functionalities.
After expanding the seed to a full family and a clean family, the protein sequences in each full or clean family are aligned using Muscle version 3.6 (23). The alignment is then filtered to retain only conserved regions, as described in Li et al. (4). For TreeFam v1, the filtered alignment was used as input in a neighbour-joining (NJ) algorithm, which was used to construct a phylogenetic tree based on amino acid mismatch distances. Since TreeFam v1, we have greatly refined our tree-building process so that the automatic trees are substantially more accurate (24). We describe the improvements to the tree building method used in TreeFam-4 subsequently.
For TreeFam v4, for each B ‘clean’ family five trees were built:
a maximum likelihood (ML) tree built using phyml (25), based on the protein alignment with the WAG model;
an ML tree built using phyml, based on the codon alignment with the HKY model;
an NJ tree using p-distance, based on the codon alignment;
an NJ tree using dN distance, based on the codon alignment; and
an NJ tree using dS distance, based on the codon alignment.
For (i) and (ii), we used a modified version of phyml release 2.4.5 (Heng Li, unpublished manuscript) which takes an input species tree, and tries to build a gene tree that is consistent with the topology of the species tree. This ‘species-guided’ phyml uses the original phyml tree-search algorithm (25). However, the objective function maximized during the tree-search is multiplied by an extra likelihood factor not found in the original phyml. This extra likelihood factor reflects the number of duplications and losses inferred in a gene tree, given the topology of the species tree. The species-guided phyml allows the gene tree to have a topology that is inconsistent with the species tree if the alignment strongly supports this. The species tree was based on the NCBI taxonomy tree (see ‘Orthologue Inference’ section subsequently).
The final tree for a B clean family is made by merging the five trees into one consensus tree using a novel ‘tree merging’ algorithm (24). This allows us to take advantage of the fact that DNA-based trees often are more accurate for closely related parts of trees and protein-based trees for distant relationships, and that some algorithms may outperform others under certain scenarios. The algorithm simultaneously merges the five input trees into a consensus tree. The consensus topology contains clades found in any of the input trees, where the clades chosen are those that minimize the number of duplications and losses inferred, and have the highest bootstrap support. Branch lengths are estimated for the final consensus tree based on the DNA alignment, using phyml with the HKY model.
We cannot use tree merging for the B full families, because it requires DNA sequences, which many UniProt proteins in full families lack. Instead, for each B full family we built an ML tree that was based on the protein alignment, and was constrained to be consistent with the tree for the corresponding clean family. The constrained ML tree was built using a modified version of phyml release 2.4.5 (Heng Li, unpublished manuscript) that can take the topology of an input gene tree as a soft constraint.
The species-guided version of phyml, the ‘constrained phyml’, and the tree merging algorithm are available as part of the TreeBest software from http://treesoft.sourceforge.net/.
Manually curating TreeFam-B trees
During curation, experts manually correct errors in the automatic trees for TreeFam-B families (4). Since TreeFam v1, significant improvements to allow curation of larger trees and to speed up curation have been made to one of our in-house curation tools, tctool (Lachlan Coin, manuscript in preparation).
TreeFam is now able to support external curation from outside the Sanger Institute, and this is currently in testing with a number of groups who are collaborating on the TreeFam project. We have recruited and trained external curators at the University of Southern Denmark in Odense, University of Aarhus and the Beijing Genomics Institute, who have contributed many curations to TreeFam.
Maintaining TreeFam-A
When a B tree has been curated, it becomes the seed tree for an A family, and is removed from TreeFam-B. Each seed family is expanded into a full and a clean family. If a new gene prediction set has been released since the last build of the TreeFam-A database, blast and hmmer are used to identify sequence matches in this gene set, which are added to the clean and/or full family. A filtered alignment is made for each full or clean family.
Trees of clean A families are built by using the tree merging algorithm to find the consensus of seven trees:
a constrained ML tree built using phyml, based on the protein alignment with the WAG model;
a constrained ML tree built using phyml, based on the codon alignment with the HKY model;
an unconstrained NJ tree using p-distance, based on the codon alignment;
an unconstrained NJ tree using dN distance, based on the codon alignment;
an unconstrained NJ tree using dS distance, based on the codon alignment;
a constrained NJ tree using dN distance based on the codon alignment; and
a constrained NJ tree using p-distance based on the codon alignment.
Trees (i) and (ii) were built using ‘species-guided PHYML’, using the topologies of the curated seed tree and of an input species tree as soft constraints. Trees (vi) and (vii) were built using the ‘constrained NJ algorithm’ described in Li et al. (4), which uses the topology of the curated seed tree as a hard constraint.
For each full A family we used constrained phyml to build a ML tree based on the protein alignment, constraining the tree to be consistent with that for the corresponding clean family.
Orthologue inference
For both A and B families, orthologues and paralogues are inferred from the clean tree. We first use the ‘Duplication/Loss Inference’ (DLI) algorithm (4,24) to identify duplication and speciation nodes. We then assume that genes belonging to different child clades of a duplication node are paralogues, while genes belonging to different child clades of a speciation node are orthologues.
Since TreeFam v1, we have introduced one change in the way that we infer orthologues, as follows. We infer that a duplication node is ‘dubious’ if there is no intersection between the species that belong to its two-child clades. A ‘dubious duplication’ is probably a tree-building artefact, and we assume that the genes belonging to the different child clades of the node are actually orthologues (not paralogues).
The DLI algorithm requires a species tree, and for this we use the NCBI taxonomy tree (7), with two exceptions. We consider two parts of the tree as multifurcations because their topology is controversial: (i) the fungi, metazoans and plants and (ii) the chordates, arthropods, nematodes and schistosomes.
TreeFam database content
Release 4 of TreeFam contains curated trees for 1314 families and automatically generated trees for another 14 351 families. The number of curated families has increased since TreeFam v1, which contained 690 curated families. The 15 665 families represent 348 531 genes from 25 fully sequenced animal genomes and 78 209 genes from four outgroups and UniProt. TreeFam v4 includes 84.5% of the 22 855 protein-coding human genes, 84.8% of the 24 438 mouse genes, 71.6% of the 14 039 D. melanogaster genes and 66.2% of the 20 060 genes from C. elegans. Table 1 shows the numbers of genes and human orthologues for each fully sequenced species in TreeFam v4.
Table 1.
The number of genes from each fully sequenced animal species that have human orthologues in TreeFam
| Species | Number of genes | Number of genes with human orthologues | Species | Number of genes | Number of genes with human orthologues |
|---|---|---|---|---|---|
| Human | 22 855 | – | C. intestinalis | 14 278 | 6189 |
| Chimpanzee | 20 982 | 18 247 | C. savignyi | 11 717 | 5215 |
| Macaque | 22 045 | 17 609 | D. melanogaster | 14 039 | 6948 |
| Mouse | 24 438 | 17 827 | D. pseudoobscura | 9871 | 5461 |
| Rat | 23 299 | 17 681 | Aedes | 31 958 | 7103 |
| Cow | 23 231 | 16 501 | Anopheles | 13 277 | 5843 |
| Dog | 18 214 | 15 546 | Flatworm | 12 799 | 4163 |
| Opossum | 19 597 | 15 973 | C. elegans | 20 060 | 5255 |
| Chicken | 18 632 | 11 973 | C. briggsae | 19 528 | 5914 |
| Frog | 18 473 | 12 018 | C. remanei | 25 555 | 6365 |
| Takifugu | 22 008 | 14 302 | Baker's yeast | 6680 | 2166 |
| Tetraodon | 28 005 | 14 246 | Fission yeast | 5043 | 2230 |
| Zebrafish | 24 948 | 16 675 | Rice | 41 252 | 6644 |
| Medaka | 20 961 | 13 804 | Thale cress | 26 207 | 6654 |
| Stickleback | 20 880 | 14 841 |
Using TreeFam
TreeFam allows users to search for their genes of interest using accession numbers from the source sequence databases or GenBank accessions, or text searches of the gene and TreeFam family names, symbols and descriptions. Since TreeFam v1, we have added the ability for users to search using GO term identifiers, Pfam domain identifiers and identifiers from many other databases (the complete list can be found at http://www.treefam.org/cgi-bin/misc_page.pl?faq#u1).
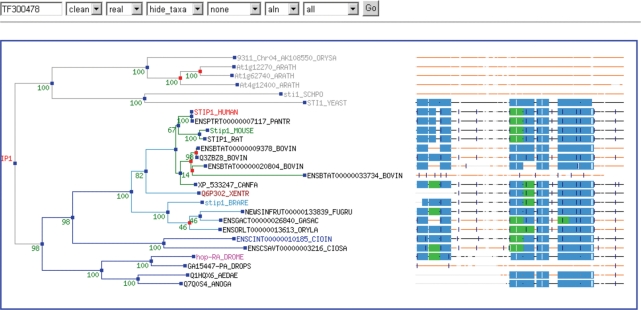
The webpage for a B family displays the clean tree, while the webpage for an A family displays both the clean and curated seed tree. Since TreeFam v1, we have added a link from the family page to the TreeView applet (Figure 1), with which users can view the full, clean or seed tree. Next to the phylogenetic tree, TreeView displays Pfam protein domains and intron positions in the family members, mapped onto the family protein alignment. The user can click on a gene name in TreeView to see the hmmer score for the match between the gene and the family.
Figure 1.
Screenshot of the TreeView applet.
All the data can be freely downloaded from ftp://ftp.sanger.ac.uk/pub/treefam. This includes sequences, alignments, trees, orthologues and within-species paralogues.
Since TreeFam v1, we have made the mysql database publicly accessible (URI: db.treefam.org Port: 3308, with user ‘anonymous’). We have also developed a perl API for interacting with the database, which allows users to fetch alignments and manipulate trees. The API and examples of using it are found at http://treesoft.sourceforge.net/.
Since TreeFam v1 we have helped the developers of the UCSC browser (26), WormBase (16) and HGNC (27) to add links to TreeFam and TreeFam orthologue information to their databases.
DISCUSSION AND FUTURE PLANS
TreeFam aims to define a gene family as a group of genes that descended from a single gene in the last common ancestor (LCA) of all animals, or that first appeared within the animals. Our methods used for grouping genes into families generally obey this rule. Unfortunately, there are exceptions: some TreeFam families contain descendants of two animal LCA genes, and the descendants of some animal LCA genes are split into two families. In addition, some TreeFam families are missing genes, or contain genes they should not.
In TreeFam v4, we have investigated using a clustering technique called ‘hcluster_sg’ to group genes into families (Heng Li, unpublished manuscript). This clusters all genes into families by using hierarchical clustering based on all-versus-all blast scores. We call the resultant families ‘TreeFam-C’ families. We are developing an algorithm to reconcile the differences between C families and the corresponding B (or A) families, and intend to use TreeFam-C to expand the gene coverage of TreeFam-B and -A.
Since TreeFam v1, TreeFam and the Compara group from Ensembl have been collaborating to converge on methods for identifying families, building phylogenetic trees and inferring orthologues and paralogues. Ensembl-Compara is focused on vertebrate genomes, while TreeFam spans the whole animal kingdom. Since Ensembl release 42 (December 2006), Ensembl-Compara has been using the tree merging algorithm used to build B clean trees in TreeFam v4. Ensembl-Compara and TreeFam also use the same algorithm for inferring orthologues and paralogues, and the same species tree. We are aiming in the future to have consistent membership of gene families between TreeFam and Ensembl.
We are also developing a service to allow users to submit a DNA or protein sequence to TreeFam, and be returned a phylogenetic tree of their gene and the members of the family to which it is most closely related.
ACKNOWLEDGEMENTS
This project is supported by The Wellcome Trust, the Chinese Academy of Science (GJHZ0701-6; KSCX2-YW-N-023), the National Natural Science Foundation of China (90403130; 90608010; 30221004; 90612019; 30725008), the Chinese 973 program (2007CB815701; 2007CB815703; 2007CB815705), the Chinese 863 program (2006AA02Z334; 2006AA10A121), the Chinese Municipal Science and Technology Commission (D07030200740000), the Chinese Ministry of Education (XXBKYHT2006001), the Danish Platform for Integrative Biology, the Ole Rømer grant from the Danish Natural Science Research Council and a pig bioinformatics grant from Danish Research Council. J.-K.H. is supported by the European Union Integrated Project MitoCheck (LSHG-CT-2004-503464). A.C. is supported by an EMBO Long-Term Fellowship. Funding to pay the Open Access publication charges for this article was provided by provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 2.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 3.Brown D, Sjolander K. Functional classification using phylogenomic inference. PLoS Comput. Biol. 2006;2:e77. doi: 10.1371/journal.pcbi.0020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Coghlan A, Ruan J, Coin LJ, Heriche JK, Osmotherly L, Li R, Liu T, Zhang Z, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen F, Mackey AJ, Stoeckert C.J., Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnamurthy N, Brown DP, Kirshner D, Sjolander K. PhyloFacts: an online structural phylogenomic encyclopedia for protein functional and structural classification. Genome. Biol. 2006;7:R83. doi: 10.1186/gb-2006-7-9-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dehal PS, Boore JL. A phylogenomic gene cluster resource: the Phylogenetically Inferred Groups (PhIGs) database. BMC Bioinformatics. 2006;7:201. doi: 10.1186/1471-2105-7-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meinel T, Krause A, Luz H, Vingron M, Staub E. The SYSTERS Protein Family Database in 2005. Nucleic Acids Res. 2005;33:D226–D229. doi: 10.1093/nar/gki030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dufayard JF, Duret L, Penel S, Gouy M, Rechenmann F, Perriere G. Tree pattern matching in phylogenetic trees: automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics. 2005;21:2596–2603. doi: 10.1093/bioinformatics/bti325. [DOI] [PubMed] [Google Scholar]
- 14.Huerta-Cepas J, Dopazo H, Dopazo J, Gabaldon T. The human phylome. Genome. Biol. 2007;8:R109. doi: 10.1186/gb-2007-8-6-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bieri T, Blasiar D, Ozersky P, Antoshechkin I, Bastiani C, Canaran P, Chan J, Chen N, Chen WJ, et al. WormBase: new content and better access. Nucleic Acids Res. 2007;35:D506–D510. doi: 10.1093/nar/gkl818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM. FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas BJ, Wortman JR, Ronning CM, Hannick LI, Smith R.K., Jr, Maiti R, Chan AP, Yu C, Farzad M, et al. Complete reannotation of the Arabidopsis genome: methods, tools, protocols and the final release. BMC Biol. 2005;3:7. doi: 10.1186/1741-7007-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 21.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas BJ, Berriman M, Hirai H, Cerqueira GG, Loverde PT, El-Sayed NM. Schistosoma mansoni genome: Closing in on a final gene set. Exp. Parasitol. 2007;117:225–228. doi: 10.1016/j.exppara.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H. Constructing the TreeFam database. 2006. PhD thesis, The Institute of Theoretical Physics, Chinese Academy of Science. http://www.sanger.ac.uk/Users/lh3/PhD-thesis-liheng-2006-English.pdf.
- 25.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Povey S, Lovering R, Bruford E, Wright M, Lush M, Wain H. The HUGO Gene Nomenclature Committee (HGNC) Hum. Genet. 2001;109:678–680. doi: 10.1007/s00439-001-0615-0. [DOI] [PubMed] [Google Scholar]