Abstract
The molecular basis of drug action is often not well understood. This is partly because the very abundant and diverse information generated in the past decades on drugs is hidden in millions of medical articles or textbooks. Therefore, we developed a one-stop data warehouse, SuperTarget that integrates drug-related information about medical indication areas, adverse drug effects, drug metabolization, pathways and Gene Ontology terms of the target proteins. An easy-to-use query interface enables the user to pose complex queries, for example to find drugs that target a certain pathway, interacting drugs that are metabolized by the same cytochrome P450 or drugs that target the same protein but are metabolized by different enzymes. Furthermore, we provide tools for 2D drug screening and sequence comparison of the targets. The database contains more than 2500 target proteins, which are annotated with about 7300 relations to 1500 drugs; the vast majority of entries have pointers to the respective literature source. A subset of these drugs has been annotated with additional binding information and indirect interactions and is available as a separate resource called Matador. SuperTarget and Matador are available at http://insilico.charite.de/supertarget and http://matador.embl.de
INTRODUCTION
Within the past two decades our knowledge about drugs, their mechanisms of action and target proteins has increased rapidly. Nevertheless, knowledge on their molecular effects is far from complete. For some drugs even the primary targets are still unknown, for example, Diloxanide, Niclosamide and Ambroxol are administered successfully although their effect on human metabolism is still not clarified at a molecular level (1). Even if the medical effect has been explained by a certain molecular interaction, most drugs interact with several additional targets, which may either strengthen the therapeutic effect or cause unwanted adverse drug effects (2). Moreover, our knowledge on drugs and their targets is highly fragmented, most of it residing in millions of medical articles and textbooks, which precludes systematic studies.
Several databases exist that collect binding data on small molecules, in particular drugs and proteins. The largest such resource is DrugBank (3), which contains 2600 drug-target relations for 900 FDA-approved drugs and additional annotations for 3200 experimental drugs. Another notable database is the Therapeutic Target Database (TTD) (4), which holds target information on about 1000 small molecule drugs. Unfortunately, DrugBank only provides references on the target, although generally not on the interactions, which makes it difficult to obtain information on the experimental context under which an interaction was observed. Moreover, the drugs in the TTD are not cross-linked with compound databases such as PubChem, ChemDB or the commercial CAS® Registry, and the targets are not linked to protein databases such as UniProt or PDB. This makes it difficult to retrieve information such as the chemical structure of the drug, its physiochemical properties, the sequence or 3D structure of its target or the biological pathways that it affects.
In order to be able to derive further information about drug-target relations, we have developed a one-stop data warehouse, SuperTarget that provides this functionality and integrates drug-target relations from different resources using heterogeneous retrieval methods. We consider a drug-target relation as a specific interaction of a small chemical compound administered to treat or diagnose a disease and a macromolecule, namely protein, DNA or RNA. The first release of SuperTarget contains a core dataset of about 7300 drug-target relations of which 4900 interactions have been subjected to a more extensive manual annotation effort to incorporate additional binding information as well as indirect interactions. The resulting data on 775 drugs is provided separately as Matador (Manually Annotated Targets And Drugs Online Resource).
DRUG-TARGET RELATIONSHIPS
Drug-target relationships described in SuperTarget were obtained in three different ways. Starting with 2400 drugs and their synonyms from the SuperDrug Database (5), the text mining tool EbiMed (6) was used to extract relevant text passages containing potential drug-target relations from about 15 millions public abstracts listed in PubMed. Many thousands of false positive or irrelevant relations were eliminated by manual curation.
In parallel, potential drug-target relations were automatically extracted from Medline by searching for synonyms of drugs, proteins and Medical Subject Headings (MeSH terms) describing groups of proteins (7). MeSH terms were used to capture and down-weight interactions that are not explicitly described in the abstracts e.g. for protein families or protein complexes. In the case of families, the specific interacting family member might not be known yet, whereas in the case of complexes, the drug might interact with more than one subunit. Proteins associated to MeSH terms were assigned by a semi-automated procedure relying on mappings provided by MeSH and synonyms of proteins that are aggregated in the STRING resource (8). Proteins that were often mentioned in abstracts, but could not be automatically assigned to families, were manually assigned. Depending on the size and nature of the families, the confidence of an interaction between drugs and individual proteins was decreased. More heterogeneous families are assigned a lower confidence. The most probable candidates were identified using a benchmarking scheme (8) and manually curated.
In a last step, relations from other databases, namely DrugBank (3), KEGG (9), PDB (10), SuperLigands (11) and TTD (4), were checked for drug-target interactions not identified with the preceding steps. If those interactions could be confirmed by literature listed in PubMed, the references were included in SuperTarget otherwise the describing database is referenced.
In consideration of the large number of entries we cannot rule out that some of the data is erroneous, change over time or is too unspecific. In the case of doubt we refer to the referenced relation source.
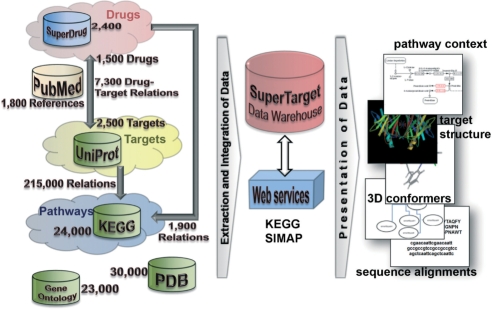
To be able to obtain more information on the drug-target relations, SuperTarget provides links to physicochemical properties and further structural information of drugs. Proven or potential target proteins are represented by sequences as stored in UniProt (12), by functional annotations extracted from GOA (13) and by related pathway information provided by KEGG (9) (compare Figure 1). Adverse drug reactions were extracted from the free accessible Canadian Adverse Reaction Monitoring Program (CADRMP, http://www.hc-sc.gc.ca/).
Figure 1.
System architecture and number of database entries of SuperTarget. The database contains the complete Uniprot with more than 3 million entries. Beside the targets, drugs and pathways the database provides 23 000 different GO-terms and 30 000 links to protein structures.
For a subset of the drug-target relations, namely those where our text-mining approach indicated a wealth of additional information, the type of binding was further analyzed and direct and indirect interactions were manually distinguished. Indirect interactions can, for example, be caused by active metabolites of the drug or by changes in the expression of a protein. The extensively annotated subset, which is contained in Matador should be well-suited as training set for various large-scale discovery approaches.
ANALYSIS OPTIONS IN SUPERTARGET
The integration of drug related information provides numerous different query entry points as well as global surveys on complex topics using heterogeneous data types (for an illustration of query capabilities see Figure 2).
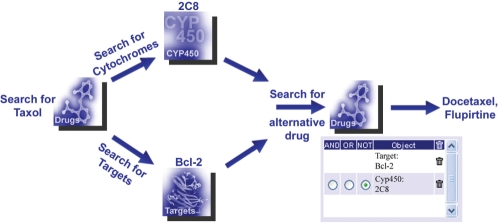
Figure 2.
Example of a complex query: search for an alternative drug to Taxol, which adresses the same target Bcl-2, but is not metabolized by the cytochrome 2C8. Procedure: start to search for the targets and the metabolizing cytochromes of Taxol in the associated categories. The resulting lists contain among others the target Bcl-2 as well as the P450 cytochrome 2C8. Second, forward 2C8 and Bcl-2 into the query term box, combine them using the ‘NOT’ operator and submit the query. The resulting drug list contains two alternative drug candidates for Taxol (Docetaxel and Flupirtine).
Since compounds with high structural similarity frequently exhibit similar activities (14), all drugs with comparable structural fingerprints are made accessible. Fingerprints allow a fast identification of drugs with very similar physiochemical properties that may interact with the same target molecules. Structural similarity scores and fingerprints are computed with the open-source Chemistry Development Kit (15).
Analogously, similar proteins are quickly identifiable by precomputed sequence alignments provided by SIMAP (16). Proteins homologous to annotated target proteins are candidates for interacting with the drug and may explain adverse effects of drugs.
Drug metabolizing enzymes come into question to explain adverse drug responses. Genetic polymorphism of cytochrome P450 genes or associated regulatory factors may lead to diverse ability of drug degradation (17). The web interface provides an extra section to retrieve the cytochrome interaction data for most of the annotated drugs.
Drugs are classified in the Anatomical Therapeutic Chemical Classification System (ATC). This hierarchical classification system was introduced by the World Health Organization in 1990, classifying drugs according to their medical indications and chemical scaffold. Groups of drugs can be selected by their ATC code. A searchable, hierarchical ATC-tree enables a fast detection of drug classes and medical indications like, for example, ‘anti-Parkinson drugs’ by expanding the branch ‘nervous system’.
The integration of Gene Ontology (GO) terms in SuperTarget enables complex queries like ‘Are there anti-cancer drugs that induce apoptosis and are associated with transcription factors?’ which can be answered by combined selection of apoptosis in the pathway tab and transcriptional activator activity in the ontology tree.
To facilitate the analysis, targets or drugs can be collected and stored in a clipboard. A session can be saved on the server and restored up to 30 days later. Each object of the clipboard links to a window with object-related information. Depending of the object type, the window contains additional information relating to drugs, targets, pathways, GO terms or metabolization. A further hyperlink leads to the search history.
CONCLUSIONS AD FUTURE DIRECTIONS
Although the first version of SuperTarget with all the search and discovery tools around drug-target relations is already an extensive resource for both large-scale research and in depth analysis, the captured knowledge is still far from complete and we would like to invite the community to help in increasing quality and quantity of the records. SuperTarget offers an option to upload and incorporate drug-target relations into a working queue. Uploaded entries will be reviewed and approved by an annotation team comprised of graduated scientists. Both SuperTarget and Matador can be used as knowledge sources, discovery tools or training sets for various applications in chemical biology and elsewhere.
AVAILABILITY
SuperTarget and Matador are available via their web sites, http://insilico.charite.de/supertarget and http://matador.embl.de. They can be obtained via a Creative Commons Attribution-Noncommercial-Share Alike 3.0 License.
ACKNOWLEDGEMENTS
The authors wish to thank B. Grüning, P. Pyl, D. Jansen, J. Tschörtner, M. Füllbeck, E. Michalsky, J. Hossbach and I. Jaeger for assistance during literature screening, software testing and improvement of the web interface. Furthermore, we thank M. Kanehisa for providing the KEGG web service as well as R. Arnold for the support using the SIMAP web service. This work was supported by BMBF (Quantpro), Deutsche Forschungsgemeinschaft (DFG: SFB 449), IRTG Berlin-Boston-Kyoto, Investitionsbank Berlin (IBB) and Deutsche Krebshilfe. Funding to pay the Open Access publication charges for this article was provided by EMBL.
Conflict of interest statement. None declared.
REFERENCES
- 1.Imming P., Sinning C., Meyer A. Drugs, their targets and the nature and number of drug targets. Nat. Rev. Drug Discov. 2006;5:821–834. doi: 10.1038/nrd2132. [DOI] [PubMed] [Google Scholar]
- 2.Frantz S. Drug discovery: playing dirty. Nature. 2005;437:942–943. doi: 10.1038/437942a. [DOI] [PubMed] [Google Scholar]
- 3.Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., Stothard P., Chang Z., Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X., Ji Z.L., Chen Y.Z. TTD: Therapeutic Target Database. Nucleic Acids Res. 2002;30:412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goede A., Dunkel M., Mester N., Frommel C., Preissner R. SuperDrug: a conformational drug database. Bioinformatics. 2005;21:1751–1753. doi: 10.1093/bioinformatics/bti295. [DOI] [PubMed] [Google Scholar]
- 6.Rebholz-Schuhmann D., Kirsch H., Arregui M., Gaudan S., Riethoven M., Stoehr P. EBIMed–text crunching to gather facts for proteins from Medline. Bioinformatics. 2007;23:e237–e244. doi: 10.1093/bioinformatics/btl302. [DOI] [PubMed] [Google Scholar]
- 7.Jensen L.J., Saric J., Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat. Rev. Genet. 2006;7:119–129. doi: 10.1038/nrg1768. [DOI] [PubMed] [Google Scholar]
- 8.von Mering C., Jensen L.J., Snel B., Hooper S.D., Krupp M., Foglierini M., Jouffre N., Huynen M.A., Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshpande N., Addess K.J., Bluhm W.F., Merino-Ott J.C., Townsend-Merino W., Zhang Q., Knezevich C., Xie L., Chen L., et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michalsky E., Dunkel M., Goede A., Preissner R. SuperLigands—a database of ligand structures derived from the Protein Data Bank. BMC Bioinform. 2005;6:122. doi: 10.1186/1471-2105-6-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.niProt-Consortium1 The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camon E., Magrane M., Barrell D., Lee V., Dimmer E., Maslen J., Binns D., Harte N., Lopez R., et al. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin Y.C., Kofron J.L., Traphagen L.M. Do structurally similar molecules have similar biological activity? J. Med. Chem. 2002;45:4350–4358. doi: 10.1021/jm020155c. [DOI] [PubMed] [Google Scholar]
- 15.Steinbeck C., Hoppe C., Kuhn S., Floris M., Guha R., Willighagen E.L. Recent developments of the chemistry development kit (CDK)—an open-source java library for chemo- and bioinformatics. Curr. Pharm. Des. 2006;12:2111–2120. doi: 10.2174/138161206777585274. [DOI] [PubMed] [Google Scholar]
- 16.Rattei T., Arnold R., Tischler P., Lindner D., Stumpflen V., Mewes H.W. SIMAP: the similarity matrix of proteins. Nucleic Acids Res. 2006;34:D252–D256. doi: 10.1093/nar/gkj106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita K. Cytochrome P450 and anticancer drugs. Curr. Drug Metab. 2006;7:23–37. doi: 10.2174/138920006774832587. [DOI] [PubMed] [Google Scholar]