Abstract
To organize data resulting from the phenotypic characterization of a library of 30 000 T-DNA enhancer trap (ET) insertion lines of rice (Oryza sativa L cv. Nipponbare), we developed the Oryza Tag Line (OTL) database (http://urgi.versailles.inra.fr/OryzaTagLine/). OTL structure facilitates forward genetic search for specific phenotypes, putatively resulting from gene disruption, and/or for GUSA or GFP reporter gene expression patterns, reflecting ET-mediated endogenous gene detection. In the latest version, OTL gathers the detailed morpho-physiological alterations observed during field evaluation and specific screens in a first set of 13 928 lines. Detection of GUS or GFP activity in specific organ/tissues in a subset of the library is also provided. Search in OTL can be achieved through trait ontology category, organ and/or developmental stage, keywords, expression of reporter gene in specific organ/tissue as well as line identification number. OTL now contains the description of 9721 mutant phenotypic traits observed in 2636 lines and 1234 GUS or GFP expression patterns. Each insertion line is documented through a generic passport data including production records, seed stocks and FST information. 8004 and 6101 of the 13 928 lines are characterized by at least one T-DNA and one Tos17 FST, respectively that OTL links to the rice genome browser OryGenesDB.
INTRODUCTION
To contribute to the international effort of functional analysis of the rice genome, we produced in the frame of the genomics initiative Génoplante, a library of 30 000 T-DNA insertion lines in the reference japonica cultivar Nipponbare (1). Each line harbors an average of 2.2 copies of the T-DNA residing at 1.5 locus. Aside gene disruption resulting from T-DNA integration, the equipment of the T-DNA with either a GUSA or a GAL4:GFP (2) enhancer trap system allows detection of nearby gene enhancer elements by observation of GUS activity or GFP fluorescence. Besides, the tissue culture procedure of the transformation protocol induces the transposition of the Tos17 retroelement (3) and the reinsertion of an average of 3.4 new copies of the retroelement per line in the T-DNA library. Large scale characterization of FSTs corresponding to T-DNA and Tos17 insertion sites has been recently accomplished (4,5).
In the last 5 years, a large collaborative effort of characterization and evaluation of the T-DNA rice insertion line collection has generated a wealth of phenotypic information. This effort includes the field evaluation and description for morpho-physiological traits of T1 progenies of 13 928 insertion lines, about half of these lines had been specifically screened for grain phenotypes and/or response to Magnaporthe grisea infection (detailed results are to be reported elsewhere), and the detection of GUSA or GFP activity in specific organs/cell types (2). To organize that information and make it accessible through the World Wide Web to the international community, we developed the Oryza Tag Line (OTL) database which is accessible at http://urgi.versailles.inra.fr/OryzaTagLine/. Dedicated to forward genetics searches, OTL provides a direct link to a rice reverse genetics database, the genome browser navigator OryGenesDB (6), through FST information of T-DNA and Tos17 insertion sites. Each insertion line is documented with both textual and pictorial information contained in a generic passport data that also includes production records, seed stocks, FST information, observation and segregation of morpho-physiological alterations during field evaluation and specific screens and detection of GUS or GFP activity in specific organ/tissues. Specific interfaces have been developed to facilitate forward genetic search either by keyword, trait ontology and developmental stage, referenced mutant type or organ/cell type and strength of reporter gene expression.
MATERIALS AND METHODS
Design and implementation
Oryza Tag Line has been implemented in the Relational Database Management System ORACLE v8i. The database architecture was built with a physical model deriving from an object-oriented view using the Unified Modeling Language (http://www.uml.org). All the web consultation interfaces were developed with perl CGI and DBI scripts for dynamic HTML pages production and databases connectivity, respectively. Perl parsers have been created to extract data from various types of files. For example, phenotype data were stored in Excel ™ formatted files while GUS/GFP data were integrate in a FileMaker database (http://www.filemaker.com/). Parsers are developed with a modular structure and are organized in several workflows. Some functions developed are (i) extract data, (ii) uniform syntax, (iii) check consistency, (iv) make the links with pictures, (v) create database indexes and input file format database.
Database content
The database currently describes 13 928 lines with readily available seed stocks. On going seed increase of the rest of the library will make this number reaching 30 000 in the near future. Besides the phenotype and gene expression data described below, some useful information is linked to each line. For instance, it is possible to visualize the detailed maps of the T-DNA constructs used to generate the lines.
Phenotype data
(i) Grain phenotypes: Examination of mature panicles bearing T1 seeds in 7187 primary transformants resulted in the observation of 251 (3.5%) phenotypes. The following alterations were observed with a decreasing frequency: aborted seeds, wrinkled, shrunken, round kernel, and deformed seeds. (ii) Response to Magnaporthe grisea: T1 seedlings of 4462 primary transformants were inoculated at the 4–5 leaf stage with a Magnaporthe grisea spore preparation in a phytotron and scored after 5 days for enhanced disease susceptibility or enhanced disease resistance compared to control Nipponbare. This led to the observation of 44 (1%) lines exhibiting an enhanced or decreased disease susceptibility and of 69 (1.5%) lesion mimics lines, with phenotype confirmed at least once. (iii) Field evaluation: Morpho-physiological traits were scored among 10–25 T1 progenies grown together with regularly interspaced control Nipponbare lines under agronomical conditions in a dedicated experimental field at the International Center for Tropical Agriculture (CIAT, Colombia). Phenotypic traits were scored at several growth stages. Evaluation is performed first 45 days after germination, then at flowering stage and eventually at the full grain maturity stage. Data were first stored in a working database, then integrated in OTL. Overall, 258 semi quantitative phenotypic descriptors have been scored and belong to 6 major classes including morphology, panicle traits, phenology, phyllotaxy, physiology and pigmentation. These phenotypic descriptors are also linked to corresponding Trait Ontology IDs (Table 1).
Table 1.
Distribution of the most frequent phenotypic alterations according to Trait Ontology codes
| Trait code | Term name | Percentage of the observed phenotypes (%) |
|---|---|---|
| TO:0000207 | Plant height | 15.61 |
| TO:0000436 | Spikelet sterility | 14.02 |
| TO:0000464 | Albino plantlet | 7.56 |
| TO:0000326 | Leaf color | 7.20 |
| TO:0000069 | Variegated leaf | 5.44 |
| TO:0000344 | Days to flower | 3.46 |
| TO:0000492 | Leaf shape | 3.25 |
| TO:0000567 | Tiller angle | 2.95 |
| TO:0000346 | Tiller number | 2.72 |
| TO:0000063 | Mimic response | 2.23 |
| TO:0000370 | Leaf width | 2.15 |
| TO:0000657 | Spikelet anatomy and morphology trait | 2.12 |
| TO:0000206 | Leaf angle | 2.06 |
| TO:0000072 | Awn length | 1.78 |
| TO:0000124 | Flag leaf angle | 1.72 |
| TO:0000050 | Inflorescence branching | 1.26 |
| TO:0000070 | Variegated leaf necrosis | 1.11 |
| TO:0000198 | Rootless | 0.96 |
| TO:0000019 | Seedling height | 0.87 |
| TO:0000240 | Sterile lemma length | 0.82 |
| Other | 20.71 |
In rice, like in Arabidopsis, the user should be aware that only a low frequency (5–10%) of the observed phenotypes are linked to the mutagen (T-DNA and Tos17). This is confirmed by previous reports on Tos17 (7) and on going, unpublished work on T-DNA lines in various laboratories. Regarding the OTL library, our work based on 69 lesion mimics mutants and 42 lines exhibiting enhanced or decreased response to the rice blast fungus Magnaporthe grisea has shown that 10% of the observed phenotypes are tagged by the T-DNA.
GUSA and GFP expression assay
GUS and GFP expression assays were carried out as described in (4) and (2), respectively. For GUS activity assays, we typically surveyed leaf blade, leaf sheath and flower tissues of the T0 plant, and half mature T1 seeds. Leaf blade, leaf sheath and flower tissues of T0 plants, half mature T1 seeds and root and shoot of 5 T1 seedlings, 3 and 5 days following germination were assayed for GFP detection.
FST (Flanking Sequence Tag) information
Isolation and sequencing of regions flanking the right and left borders of T-DNA inserts (1) and the 3′LTR of new Tos17 inserts (5) was carried out systematically in the mutant collection and information related to these FSTs is accessible through a link (Figure 1E and B) to the OrygenesDB database (http://orygenesdb.cirad.fr) (6), which is cross-linked to OTL. Overall, 8004 and 6101 of the 13 928 lines are characterized by at least one T-DNA and one Tos17 FST, respectively while 74.5 (%) of the lines are characterized by at least one T-DNA or Tos17 insert. Manipulation of large number of DNA and seed samples passing through PCR and field increase respectively, even assisted by a barcode system and robotics, are prone to errors like mislabelling or PCR contamination that one tries to limit as much as possible. In the case of OTL, test production of new FSTs in T2 progenies (the generation distributed to users) of 31 lines and Southern observation of locus rearrangement in T2 progenies of 60 lines have shown that the frequency of errors is low (0 and 6% respectively).
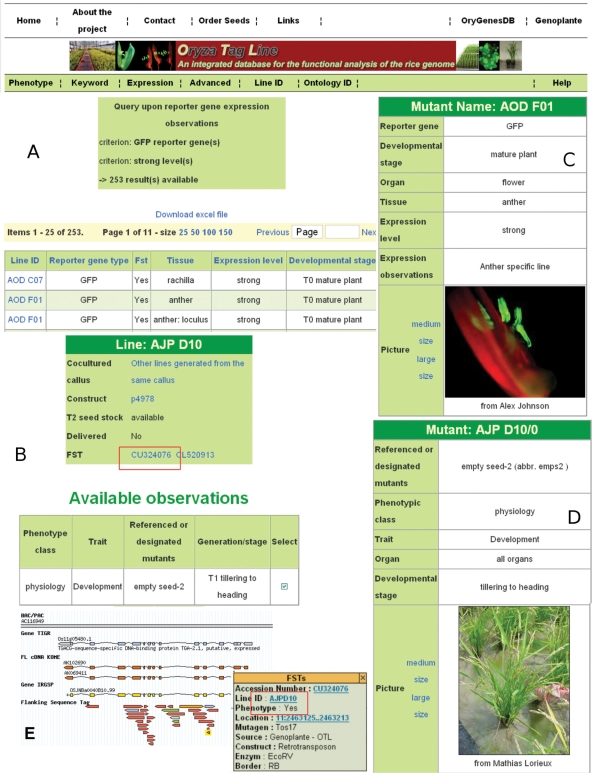
Figure 1.
Description of a mutant search. (A) Search interfaces return a list of lines. The results table displays the Line ID, the reporter gene type, the presence or absence of FST information in the Gbrowse navigators (such as OryGenesDB), the tissue where the reporter gene is expressed, the level of expression and the developmental stage of the observed plants. Details about a specific Line ID are displayed by an interactive link shown in B. (B) This interface displays all information available for a specific Line ID (i.e. general features, phenotypic and gene expression observations). Details of the insertion site are displayed through OryGenesDB by following the links on FST ID. Observations to be viewed in detail can be selected with HTML checkboxes. (C) Here, specific organ expression and pictures of the mutant organ and/or plant are presented. Pictures of increasing sizes are also available to enhance the resolution. (D) This frame displays detailed information and picture for a specific phenotypic mutant. (E) Illustration of the forth and back links from the line ID in Oryza Tag Line (red box in part B) to the physical map position and related sequence information of the Flanking Sequence Tag in the genome navigator OryGenesDB (red box in part E).
Plant ontology
We adopted the Plant Ontology Consortium (8–11) controlled vocabularies to describe mutant phenotypes in order to be consistent with other popular plant genomic databases [e.g. Gramene (11), TAIR (12), Oryzabase (13)]. In many cases, ontologies allow cross-referencing and building links between different data sources (e.g. references, gene annotation, sequences, trait observations) (14).
Availability
OTL is accessible at http://urgi.versailles.inra.fr/OryzaTagLine/. Users can request seeds of insertion lines in filling downloadable order and MTA forms at that site and/or sending an e-mail to insertionlines.crb@cirad.fr
RESULTS
Oryza Tag Line data content
13 928 T1 progeny were evaluated under agronomical conditions in the field at a single location. OTL contains 9721 records of mutant traits, falling into 6 broadly defined phenotypic classes including morphology, panicle traits, phenology, phylotaxy, physiology and pigmentation observed in a total of 2636 lines. 19% of the lines exhibited at least one variant trait in at least one fourth of the T1 progeny plants, which is the ratio expected for homozygous mutant in a Mendelian segregation at a single locus. Plant height and spikelet sterility were the two most frequent alterations. Albinism represented 7.6% of the observed phenotypes. The most represented phenotypic alterations are shown in Table 1. When a matching trait ontology description is available, each phenotype is described with Plant Ontology Consortium (10) controlled vocabularies.
A total of 27 and 29% of the lines assayed for vegetative and floral tissues allowed detection of GUS activity or GFP fluorescence respectively in at least one of the organs scored. Overall, 1234 lines exhibited GUSA and GFP reporter gene expression patterns with several levels of description, from the organ level for all lines to confocal observations at the cell/tissue level in a few lines. 30.1, 18.2 and 25.6% of the 2636 lines exhibiting at least one mutant phenotypic trait are characterized by at least one T-DNA FST, at least one Tos17 FST and both, respectively. Taking into account an interval −1000 upstream ATG to 300 bp downstream STOP codon of predicted genes, we also estimated that 4511 and 2572 non-TE genes of rice were interrupted by T-DNA and Tos17 inserts respectively in the part of the library which has been evaluated. The T1 lines grown in the field were allowed to self-pollinate and T2 seeds of the 13 928 T2 lines are now publicly available.
Oryza Tag Line interface
Oryza Tag Line is accessible at http://urgi.versailles.inra.fr/OryzaTagLine/. Users can search information from OTL in several ways.
‘Phenotype search’: The ‘Phenotype search’ interface allows retrieving a list of selected lines exhibiting specific features. For example, a ‘Phenotype search’ can be used to identify alterations observed at a precise developmental stage (e.g. tillering) in a specific organ (e.g. leaf) and/or belonging to a particular class of traits (e.g. Morphology). Some additional criteria can be used to restrict the search only to lines with available T2 seed stock and/or with characterized insertion sites (i.e. with T-DNA and/or Tos17 FST).
‘Keyword search’: The ‘Keyword search’ is performed over all the phenotypic fields (e.g. mutant name, trait description, trait name, general observation, synonyms). It results in a powerful interface allowing searches in any field of phenotypic description. Moreover, wildcard (*) is allowed for an incomplete search or missing word.
‘Expression search’: The ‘Expression search’ interface displays lines with reporter gene expression in specific tissues or organs. The database mainly contains GAL4-mediated GFP expression data presented in (2) and unpublished GUSA reporter gene expression profiles. Additional details for specific lines are accessible thanks to a pop up menu (e.g. reporter gene, expression level, organ, tissue). Users can also use features to restrict the list of lines displayed. Search for a specific pattern is possible in combination to FST and seed stock availability as in all search interfaces.
‘Advanced search’: The ‘Advanced search’ allows complex queries combining free text search mixing phenotypic terms, plant organs and reporter gene data. For each field, a search domain has to be selected. Text search can be more precisely defined with operators like ‘contains’, ‘starts with’, ‘ends with’ and ‘exactly’. While ‘phenotype’ or ‘keywords’ search interfaces propose a single search, ‘advanced’ search interface is specifically designed to search lines with multiple phenotype observations (e.g. trait name, referenced mutant or its abbreviation, reporter gene expression, localization and level of expression).
‘Using plant ontology controlled vocabularies’: All data integrated into Oryza Tag Line are described with ontologies according to the Plant Ontology Consortium (8–11) (e.g. trait, plant structure, cereal plant growth stages). For data integrated into Oryza Tag Line, each term has been linked to many ontology ID (e.g. anatomy, trait and growth stage). For example a mutant with large ligules is described with the Trait Ontology term ‘ligule length’ (TO:0000024) has two terms from plant ontology [e.g. leaf (PO:0009025) and ligule (PO:0020105)]. However, some mutant observed such as the ‘lined up tillers’ mutant do not match any clear ontology term. This mutant is characterized by its tillers arranged in a straight line combined with dwarf (−70%) in proportion, low tillering, late flowering and an incomplete exertion of panicles phenotypes. In this case, this mutant is then temporarily classified with a more general ontology term like ‘stem anatomy and morphology trait (TO:0000361)’ but the full mutant description is still visible.
‘Retrieve list output’: A user can organize output tables so as to sort data by clicking on the headers. The output tables can be downloaded as separate Excel ™ formatted files.
CONCLUSION
With 13 928 entries related to 266 traits of interest, Oryza Tag Line represents, along with the NIAS Tos17 (15) and the RMD T-DNA (16) databases, a valuable resource for both discovering functions of novel genes underlying agronomical traits and retrieving information about alterations putatively caused by insertions in candidate sequences, in the model cereal plant, rice. New observations resulting from the continuing field evaluation of insertion lines will be integrated in OTL. We plan to add future cross references with other popular plant genomic databases [e.g. Gramene (10), Oryzabase (13)] through sharing information using MOBY network and web service technologies in the long run. Moreover, ontologies will also facilitate a common search in the expanding number of rice mutant databases [e.g. IRRI IR64 deletion mutant library: http://www.iris.irri.org/icis/SearchIRIS.htm (17), RMD: http://rmd.ncpgr.cn/(16), NIAS Nipponbare retrotransposon insertion library: http://pc7080.abr.affrc.go.jp/%7Emiyao/pub/tos17 (15)] through a common portal which is under development at the International Rice Functional Genomics Consortium (IRFGC, http://www.iris.irri.org/IRFGC/default.shtml).
ACKNOWLEDGEMENTS
This work was supported by the French Genomics Initiative Génoplante. The authors would like to thank technical assistance which has contributed to the generation of the biological data integrated into Oryza Tag Line. The authors thank Pietro Piffanelli and Christine Tranchant for their help during the database interface conception and evaluation; Anne Diévart for her help in writing this paper; Delphine Samson, Fabrice Legeai, Sébastien Reboux and François Artiguenave for delivering access to the database. This work was supported by the genomics initiative Génoplante and benefited of the Infrastructures of the Génopole Montpellier LR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sallaud C, Gay C, Larmande P, Bes M, Piffanelli P, Piegu B, Droc G, Regad F, Bourgeois E, et al. High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J. 2004;39:450–464. doi: 10.1111/j.1365-313X.2004.02145.x. [DOI] [PubMed] [Google Scholar]
- 2.Johnson A.AT, Hibberd JM, Gay C, Essah PA, Haseloff J, Tester M, Guiderdoni E. Spatial control of transgene expression in rice (Oryza sativa L.) using the GAL4 enhancer trapping system. Plant J. 2005;41:779–789. doi: 10.1111/j.1365-313X.2005.02339.x. [DOI] [PubMed] [Google Scholar]
- 3.Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. PNAS. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sallaud C, Meynard D, van Boxtel J, Gay C, Bes M, Brizard JP, Larmande P, Ortega D, Raynal M, et al. Highly efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theor. Appl. Genet. 2003;106:1396–1408. doi: 10.1007/s00122-002-1184-x. [DOI] [PubMed] [Google Scholar]
- 5.Piffanelli P, Droc G, Mieulet D, Lanau N, Bès M, Bourgeois E, Rouvière C, Gavory F, et al. Large-scale characterization of Tos17 insertion sites in a rice T-DNA mutant library. plant Mol. Biol. doi: 10.1007/s11103-007-9222-3. in press. doi: 10.1007/s11103-007-9222-3. [DOI] [PubMed] [Google Scholar]
- 6.Droc G, Ruiz M, Larmande P, Pereira A, Piffanelli P, Morel JB, Dievart A, Courtois B, Guiderdoni E, et al. OryGenesDB: a database for rice reverse genetics. Nucleic Acids Res. 2006;34:D736–D740. doi: 10.1093/nar/gkj012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirochika H. Contribution of the Tos17 retrotransposon to rice functional genomics. Curr. Opin. Plant Biol. 2001;4:118–122. doi: 10.1016/s1369-5266(00)00146-1. [DOI] [PubMed] [Google Scholar]
- 8.Ilic K, Kellogg EA, Jaiswal P, Zapata F, Stevens PF, Vincent LP, Avraham S, Reiser L, Pujar A, et al. The plant structure ontology, a unified vocabulary of anatomy and morphology of a flowering plant. Plant Physiol. 2007;143:587–599. doi: 10.1104/pp.106.092825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pujar A, Jaiswal P, Kellogg EA, Ilic K, Vincent L, Avraham S, Stevens P, Zapata F, Reiser L, et al. Whole-plant growth stage ontology for angiosperms and its application in plant biology. Plant Physiol. 2006;142:414–428. doi: 10.1104/pp.106.085720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pankaj Jaiswal SA, Katica Ilic, Elizabeth A Kellogg, Susan McCouch, Anuradha Pujar, Leonore Reiser, Seung Y Rhee, Martin M. Sachs, Mary Schaeffer, Lincoln Stein, Peter Stevens, Leszek Vincent, Doreen Ware, Felipe Zapata Plant Ontology (PO): a controlled vocabulary of plant structures and growth stages. Comparative and Functional Genomics. 2005;6:388–397. doi: 10.1002/cfg.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaiswal P, Ni J, Yap I, Ware D, Spooner W, Youens-Clark K, Ren L, Liang C, Zhao W, et al. Gramene: a bird's eye view of cereal genomes. Nucleic Acids Res. 2006;34:D717–D723. doi: 10.1093/nar/gkj154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurata N, Yamazaki Y. Oryzabase. An integrated biological and genome information database for rice. Plant Physiol. 2006;140:12–17. doi: 10.1104/pp.105.063008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camon E, Barrell D, Lee V, Dimmer E, Apweiler R. The Gene Ontology Annotation (GOA) Database—an integrated resource of GO annotations to the UniProt Knowledgebase. In Silico Biol. 2004;4:5–6. [PubMed] [Google Scholar]
- 15.Miyao A, Iwasaki Y, Kitano H, Itoh J, Maekawa M, Murata K, Yatou O, Nagato Y, Hirochika H. A large-scale collection of phenotypic data describing an insertional mutant population to facilitate functional analysis of rice genes. Plant Mol. Biol. 2007;63:625–635. doi: 10.1007/s11103-006-9118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Li C, Wu C, Xiong L, Chen G, Zhang Q, Wang S. RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Res. 2006;34:D745–D748. doi: 10.1093/nar/gkj016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruskiewich RM, Cosico AB, Eusebio W, Portugal AM, Ramos LM, Reyes MT, Sallan MA, Ulat VJ, Wang X, et al. Linking genotype to phenotype: the International Rice Information System (IRIS) Bioinformatics. 2003; 19(Suppl 1):i63–i65. doi: 10.1093/bioinformatics/btg1006. [DOI] [PubMed] [Google Scholar]