Abstract
The Genome Database for Rosaceae (GDR) is a central repository of curated and integrated genetics and genomics data of Rosaceae, an economically important family which includes apple, cherry, peach, pear, raspberry, rose and strawberry. GDR contains annotated databases of all publicly available Rosaceae ESTs, the genetically anchored peach physical map, Rosaceae genetic maps and comprehensively annotated markers and traits. The ESTs are assembled to produce unigene sets of each genus and the entire Rosaceae. Other annotations include putative function, microsatellites, open reading frames, single nucleotide polymorphisms, gene ontology terms and anchored map position where applicable. Most of the published Rosaceae genetic maps can be viewed and compared through CMap, the comparative map viewer. The peach physical map can be viewed using WebFPC/WebChrom, and also through our integrated GDR map viewer, which serves as a portal to the combined genetic, transcriptome and physical mapping information. ESTs, BACs, markers and traits can be queried by various categories and the search result sites are linked to the mapping visualization tools. GDR also provides online analysis tools such as a batch BLAST/FASTA server for the GDR datasets, a sequence assembly server and microsatellite and primer detection tools. GDR is available at http://www.rosaceae.org.
INTRODUCTION
The Genome Database for Rosaceae (GDR) is a curated and integrated web-based database containing comprehensive genetic and genomic data for the Rosaceae. Rosaceae contains a number of important fruit-producing crops: apple (Malus), pear (Pyrus), raspberries/blackberries (Rubus), strawberries (Fragaria) and stone fruits (Prunus) such as peach/nectarine, apricot, plum, cherry and almond. Rosaceae also contains a wide variety of ornamental plants including roses, flowering cherry, crabapple, quince and pear. To improve the economic competitiveness of these crops and to better understand the biological principles controlling the various traits of these plants, substantial genomic/genetic studies are underway by research groups worldwide. As a result, a wide array of genetic and genomic data for the Rosaceae species is being accumulated. Of these, ESTs (Expressed Sequence Tag) are fastest growing genomic resource. The EST sequences of cDNA libraries constructed from various tissues and in various conditions serve as a crucial tool in the development of markers for genetic mapping, the development of probes for gene expression studies or direct mapping to genomic clones or chromosomes, and in gene prediction when large genomic sequences are available. As of August 2007, there were 380 000 ESTs from 177 libraries and 20 Rosaceae species. The availability of genetic map data continues to accumulate rapidly. Peach, a member of Prunus, is considered to be the best genetically characterized species in Rosaceae. Numerous genetic maps exist for peach and other Prunus species, including the general Prunus TxE map (1), a highly saturated map with genetic markers and agronomically important traits. The TxE map is constructed from a cross of peach and almond and many of its markers serve as anchor points for numerous other Prunus genetic maps. This map is also used as a framework for the genetically anchored peach physical map and the peach transcriptome map. Genetic maps from other genera are also available for apple, pear, rose, raspberry and strawberry. Molecular markers have also been developed and utilized in the study of genetic diversity. Physical maps are available for peach (2) and apple (3), serving as invaluable resources for marker development, map-based cloning of genes, large-scale genome sequencing and comparative genomic analysis among Rosaceae and across plant families. The genetic and genomic resources of well-studied organisms can greatly accelerate the characterization of other closely related organisms. The anchor markers were shown to be essentially collinear among Prunus maps, and substantial collinear blocks were also detected among different genera in Rosaceae (4).
For these numerous and complex data to be efficiently utilized, a centralized database is essential to curate, analyze, and integrate the data and provide efficient interfaces for user access. To meet this need, the Genome Database for Rosaceae was initiated as previously reported (5). Here we describe the data and the functionality of GDR with particular focus on the new enhancements which include: (i) an annotated unigene data for each genus and the entire family, (ii) extensive Rosaceae maps, markers, and trait data, (iii) a complete peach physical map and associated genetic and transcriptome map data, (iv) an enhanced user-interface including various search sites and CMap for comparative map viewing, (v) new web-based tools such as the SSR and CAP3 assembly servers and (vi) enhanced community resources to facilitate communication among Rosaceae researchers.
DATABASE DESCRIPTION
GDR data and web interface
GDR data includes various Rosaceae genetics and genomics data including annotated EST sequences, markers, traits, genetic maps, peach BAC hybridization data, peach physical and transcriptome maps and journal article citations.
Users can access data through individual search sites for the various data types such as EST, Marker, Trait and BAC. Various maps can be viewed through graphical interfaces such as CMap and WebFPC/WebChrom. If users are interested in a particular species they can go to the species pages of interest. We currently have species pages for apple, pear, almond, apricot, cherry, peach, raspberry, rose and strawberry. Each page lists available data, analysis tools and funded projects for the species and provides hyperlinks to the specific sites in GDR and other internet sites. If users are interested in a specific project, such as Rosaceae EST unigene or peach physical map, they can go to the corresponding project page. Project pages provide data description, data summary and links to sites for search, download or further analysis. The major GDR data and the web interface to the data are described subsequently.
Rosaceae unigene and other annotated EST data
The GDR contains all the publicly available Rosaceae ESTs downloaded from the dbEST at NCBI (6). The routine processing in GDR occurs in three stages: sequence filtering and trimming to obtain high-quality sequences, assembly into contigs to reduce the inherent redundancy and build unigene sets and sequence annotation. The most recent unigene set was developed from 359 001 high-quality ESTs, representing 151 cDNA libraries, 17 species and 20 Plant Ontology curated tissue types. EST data are downloaded daily from NCBI, and uploaded to the database for browsing or searching. As of August 7, 2007 GDR contained 380 702 ESTs.
The unigene sets for four genera, Malus, Prunus, Fragaria, Rosa and Pyrus, were generated using CAP3 (7) with an overlap percentage parameter of 90 (-p 90). The assembled contigs and singlets for the four sets were again assembled to generate the putative unigene set for the entire Rosaceae ESTs. Our annotation procedure includes pair-wise comparison of both the filtered ESTs and the EST contig consensus sequences against the SWISS-PROT, TrEMBL and InterPro databases (8,9) using BLAST (10). The top ten matches with an expectation value less than 1e-6 are recorded for each EST and contig. These matches are used to assign Gene Ontology (11) terms that facilitate searching sequences by keywords and grouping sequences by similar function. Plant Structure Ontology (12) is also utilized to annotate the ESTs with the tissue from which the ESTs are generated.
Additional sequence annotation includes computational analysis of simple sequence repeats (SSR) and open reading frames (ORF) on both the filtered ESTs and the contigs. SSR analysis is performed using a modified version of a Perl script SSRIT (13) to select sequences that contain dinucleotide motifs occurring at least five times, trinucleotides four times and tetranucleotide or pentanucleotide three times. To examine the location of SSRs in the EST sequences in relation to the putative coding region, we use the FLIP (14) program. Using the FLIP output, we select the longest ORF as the putative coding region and report the location of SSRs in relation to the putative coding region. Primer3 (15) is also utilized to generate primers for the SSRs where possible using the default software parameters. The single nucleotide polymorphisms (SNPs) for the contigs are generated with the autoSNP (16) package using default stringencies for the genera unigene contigs.
In addition to the ESTs downloaded from NCBI, GDR also provides project-specific EST analyses in collaboration with other research groups. ESTs analyzed by the GDR team currently include almond, peach, strawberry and raspberry.
The project page, ‘Rosaceae EST unigene’, is a good starting point for an overview of the various annotated data for the genera and family unigenes. The page provides a chart with the numbers of ESTs, unigenes, contigs and singlets for each unigene set and links to the individual unigene pages. The individual unigene project page displays the aims and overall results of the project with a side bar containing links to the EST search sites, library details, protocols and downloads, putative homology, microsatellite analysis and contact and publication information. A link to the gene ontology (GO) classification is also available for the Rosaceae family assembly and is also being made available for the genera assemblies. The genera unigene assemblies are available from the original data overview page and include a page in the side bar linking to SNP analysis. Downloadable data includes batch sequence in fasta format, homology results file in Excel format and SSR/ORF/primers results in Excel format. Individual project pages are also available with the similar content as in the Rosaceae EST unigene pages.
The EST search site is for those users who are interested in a subset of ESTs. They can choose to search ESTs of the entire Rosaceae or the genus of interest by selecting the appropriate tabs. Users can also either search ESTs or contigs. In each search page, ESTs or contigs can be searched by their name(s), assembly results, sequence features such as SSR or SNP, taxonomy, tissue type and putative function including match description, match organism and GO term. Users can also perform a batch search by uploading a file with EST names. Previous unigene versions are also available for search to help those who have been using an older version in their research. A simple search can be conducted by typing a clone name or accession number, selecting ESTs that belong to a certain unigene set or containing sequence features such as SSR or SNP. The search category ‘tissue type’ lists all the tissues assigned from the Plant Ontology terms to the various cDNA libraries. The ‘putative function’ category allows users to enter descriptions, source organisms or GO terms relating to a gene of interest. The search will find genes with SWISS-PROT matches to any of these keywords. All of the search categories can be combined to suit the users’ need. The results can be downloaded in fasta format or as a tab-delimited file with SWISS-PROT homology results containing hyperlinks back into the data for each sequence retrieved.
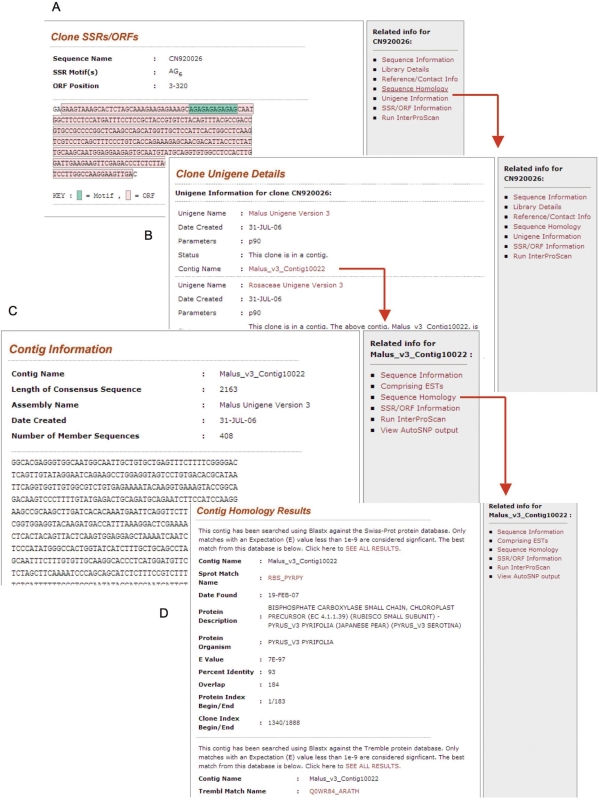
Instead of displaying all the details on one page, the EST details page initially displays the clone information and the sequence with a side bar containing links to library details, unigene information, sequence homology, SSR/ORF information, map position and anchored BACs when applicable (Figure 1). A unigene information page provides the contig name and hyperlink for both the genus and family unigenes (Figure 1). The contig page gives similar annotation data for the contig with additional links to the SNP results and the comprising ESTs. Also available from the EST or contig page is the InterProScan Web Services. This Web Service allows users to directly scan for protein signatures without going to the EBI site and uploading the sequences. For the ESTs anchored to peach BACs and/or to Rosaceae genetic maps, the EST detail page provides a link to view the ESTs' map positions using the GDR Map Viewer or CMap.
Figure 1.
EST and contig page. (A) SSR/ORF information page showing the EST sequence along with the position of SSRs and the longest ORF. (B) Unigene information page showing the assembly displaying the unigene resulted from genus or family level assembly. The unigene name is linked to the corresponding contig page. (C) Contig information page with the sequence and the side bar. (D) Contig sequence homology page showing the best SWISS-PROT and TrEMBL matches.
Rosaceae genetic map, peach physical and transcriptome map
The GDR currently contains data for 37 genetic maps from apple, pear, almond, apricot, cherry, peach, raspberry, rose, strawberry and inter-specific crosses within Prunus. We use CMap, the web-based comparative map tool, to allow users to compare maps from different cultivars and species. The comparative mapping facilitates the data transfer from well-studied species to less-studied ones. For example, the GDR map collection includes the TxE map, which is recognized as the reference map for Prunus. The TxE map, constructed from an almond × peach F2 population, contains 826 markers with a total distance of 524 cm (1,4). The TxE map contain many markers that are used in the construction of maps of other Prunus species such as peach, apricot, sour cherry, plum × almond–peach hybrid and almond x peach, but also other Rosaceae species such as apple and pear.
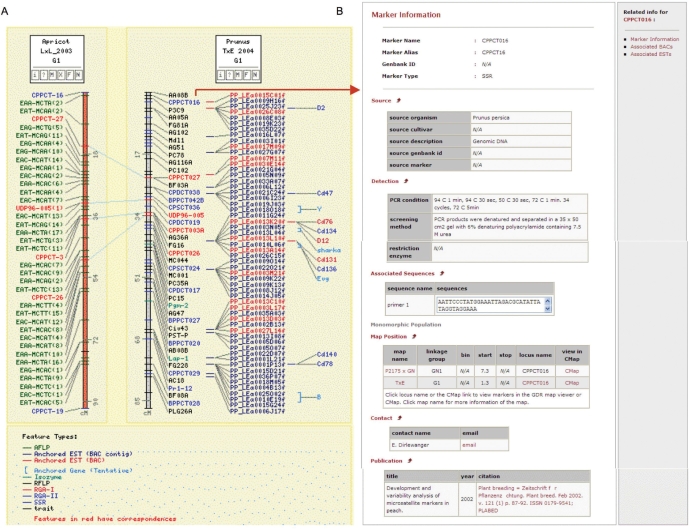
The essential collinearity of the anchored markers in the Prunus maps and the presence of large collinear blocks among different genera in Rosaceae, such as Prunus and Malus (4), enable comparative mapping, an invaluable tool for cross-utilization of data in Rosaceae. In addition to the directly-mapped genetic markers, the TxE map in GDR-CMap displays peach transcriptome map data, major trait loci affecting agronomic characters found in various Prunus species, and pathogen resistance loci (Figure 2). The peach transcriptome was established by the hybridization of the putative unique ESTs from a peach cDNA library of developing fruit mesocarp (17). The peach ESTs were anchored to the TxE map by hybridization to the genetically anchored peach BACs (17). The position of major trait loci have been established in the TxE map using the data from different linkage maps anchored with the TxE reference map (4). Candidate genes representing analogs of major resistance genes have been anchored to this map by the hybridization to the genetically anchored peach BACs (18).
Figure 2.
CMap and the marker detail page. (A) A screenshot of a CMap page that shows the comparison between G1 of TxE map and the Apricot map. The transcriptome map and the anchored trait positions are also available in TxE map. (B) A marker detail page that is hyperlinked from CMap page.
GDR-CMap serves as an integrative tool in the utilization of the data anchored to the TxE reference map in the study of other species in Prunus and other genera in Rosaceae. Genetic maps of apple and pear also share considerable markers, enabling the comparison between Malus and Pyrus. The anchored features, such as marker and ESTs, in the map are also linked to the corresponding GDR sites so that all the relevant information for the features can be viewed (Figure 2).
Another important resource in GDR is the peach physical map data. The peach physical map is constructed from two peach BAC libraries (19) using High-Information-Content Fingerprinting and FPC software. The physical length of the map is estimated to be 303 cm, which is 104.5% of the peach genome. Also available are the markers that have been used in the integration of the physical map with the peach transcriptome map and the Prunus general map. The markers include ESTs and various genetic markers that have been used in direct BAC hybridization and other genetic markers that are developed from BAC end sequences or EST-SSRs that are developed from BAC-hybridized ESTs. To date, 2636 BAC-associated markers are integrated into the physical map. GDR uses two tools available from the WebAGCoL Package (20) to display the current peach physical map. WebFPC displays the BAC contigs with associated markers and ESTs. WebChrom displays the BAC contigs anchored to each linkage group of the general Prunus map. All the ESTs, markers and BACs displayed in WebFPC or WebChrom are directly linked to the detail site of GDR. In addition, the combined genetic, physical and transcriptome map can be viewed using an in-house developed GDR map viewer. In this GDR map viewer, the general Prunus map is displayed with markers, anchored ESTs, hybridized BACs and anchored BAC contigs. Direct links to WebFPC and CMap are provided for comparative map viewing or contig assembly viewing.
Rosaceae genetic markers and traits
To provide more details of the genetic markers and traits that have been used in genetic map development or genetic diversity studies, we developed an extensively annotated molecular marker database. Currently, over 1400 extensively annotated markers are available from GDR. The marker annotation includes marker aliases, source cultivar, source description, primer sequences, PCR conditions and references, in addition to previously available data such as map position, associated ESTs and associated BACs (Figure 2). While annotation of trait data is at an initial stage, the traits are annotated in GDR with aliases, published symbol, curated trait category, taxon, trait description, screening method, map position and references. The marker search site allows both a simple search by name and an advanced search with various search categories. The search category includes marker type, the species from which the marker is developed, the species to which the marker is mapped, map position, markers with associated BAC clones and markers with associated ESTs. Users can also upload a file of names to get the detailed data. In trait search site, users can search trait by name, symbol, taxon or curated trait category.
Web-based analysis tools
GDR web-based tools include a FASTA server, BLAST server, CAP3 Assembly server and SSR server. The FASTA/BLAST servers allow users to conduct sequence homology analyses against various sequence databases including annotated sequences in GDR. The databases includes ESTs of the Rosaceae or each genus from NCBI, genera-specific unigene sets, as well as a family-wide unigene, Rosaceae genomic or protein sequences from NCBI, Arabidopsis protein sequences from TAIR and the ESTs, unigene sets, SSR-containing ESTs from individual cDNA libraries of peach mesocarp, almond, octoploid strawberry and diploid strawberry. Peach mesocarp ESTs that are anchored to the peach BACs are also available for sequence analysis. Batch sequences can be uploaded for analysis and the results are returned as both raw aligned output and parsed out in Excel. The output in Excel has hyperlinks to the GDR and NCBI sites. FASTA formatted library files of both the sequences with or without matches are provided to allow the user to easily conduct further batch searches in GDR and other databases. An EST assembly server using the CAP3 program is also available so that users can assemble their own EST sets. The server returns the raw output, a summary report and a fasta file containing the combined contig sequences and singlet sequences, which are also available as individual files. The contig file lists the contig number and comprising clone names in the comment line for each assembled transcript. Also available is a SSR server that allows user-defined SSRs to be identified in uploaded sequences. Users can also choose to run Primer3 along with the SSR-detection program to generate primer sets for the SSRs.
Community resources
GDR provides access to community-based news on various pages under the ‘community’ header bar, such as Rosaceae genomics, USRosEXEC, conferences, meetings, funding, employment, mailing lists and message boards. USRosEXEC stands for US Rosaceae Genomics, Genetics and Breeding Executive Committee, which serves as a communication and coordination focal point for the community. The USRosEXEC page provides the official documents, meeting minutes, membership and subcommittee information. Several mailing lists, in addition to the GDR mailing list, are available to serve the community with information for specific interests or purposes, and the archives can be viewed through the message board sites. All the publications in Rosaceae genomics and genetics are also available in GDR through the publication search site.
CONCLUSION AND FUTURE DIRECTION
We have substantially extended the data and the functionality of GDR since our first report (5). This is reflected in the usage of GDR, with 218 409 visits and 2 070 880 pages accessed between August 1, 2006 and July 31, 2007. The new enhancements include unigene development for the Rosaceae and each genus, Rosaceae map, marker and trait databases, the completed peach physical map, new sequence assembly and SSR servers analysis tools, and more project and community resources. We will continue in our efforts to curate and integrate future genomics and genetics data including the apple physical map, microarray data, and the forthcoming genome sequences of apple and peach.
ACKNOWLEDGEMENTS
The article was funded by National Science Foundation Plant Genome Program (#0320544 to D.M.); United States Department of Agriculture Cooperative State Research, Education and Extension Service—National Research Initiative—Plant Genome Program (#2005-35300-15452 to A.A.); Clemson University and Washington State University. Funding to pay the Open Access publication charges for the article was provided by NSF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Howad W, Yamamoto T, Dirlewanger E, Testolin R, Cosson P, Cipriani G, Monforte AJ, Georgi L, Abbott AG, et al. Mapping with a few plants: using selective mapping for microsatellite saturation of the Prunus reference map. Genetics. 2005;171:1305–1309. doi: 10.1534/genetics.105.043661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhebentyayeva TN, Horn R, Mook J, Lecouls A, Georgi L, Abbott AG, Reighard GL, Swire-Clark G, Baird WV. A physical framework for the peach genome. Acta Hort. 2006;713:83–88. [Google Scholar]
- 3.Han Y, Gasic K, Marron B, Beever JE, Korban SS. A BAC-based physical map of the apple genome. Genomics. 2007;89:630–637. doi: 10.1016/j.ygeno.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Dirlewanger E, Graziano E, Joobeur T, Garriga-Caldere F, Cosson P, Howad W, Arus P. Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc. Natl Acad. Sci. USA. 2004;101:9891–9896. doi: 10.1073/pnas.0307937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung S, Jesudurai C, Staton M, Du Z, Ficklin S, Cho I, Abbott A, Tomkins J, Main D. GDR (Genome Database for Rosaceae): integrated web resources for Rosaceae genomics and genetics research. BMC Bioinformatics. 2004;5:130. doi: 10.1186/1471-2105-5-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, et al. The universal protein resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ilic K, Kellogg EA, Jaiswal P, Zapata F, Stevens PF, Vincent LP, Avraham S, Reiser L, Pujar A, et al. The plant structure ontology, a unified vocabulary of anatomy and morphology of a flowering plant. Plant Physiol. 2007;143:587–599. doi: 10.1104/pp.106.092825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bossard N. 1997. FLIP: a Unix program used to find/translate ORFs. Bionet Software. [Google Scholar]
- 15.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 16.Barker G, Batley J, O’ Sullivan H, Edwards KJ, Edwards D. Redundancy based detection of sequence polymorphisms in expressed sequence tag data using autoSNP. Bioinformatics. 2003;19:421–422. doi: 10.1093/bioinformatics/btf881. [DOI] [PubMed] [Google Scholar]
- 17.Horn R, Lecouls AC, Callahan A, Dandekar A, Garay L, McCord P, Howad W, Chan H, Verde I, et al. Candidate gene database and transcript map for peach, a model species for fruit trees. Theor. Appl. Genet. 2005;110:1419–1428. doi: 10.1007/s00122-005-1968-x. [DOI] [PubMed] [Google Scholar]
- 18.Lalli DA, Decroocq V, Blenda AV, Schurdi-Levraud V, Garay L, Le Gall O, Damsteegt V, Reighard GL, Abbott AG. Identification and mapping of resistance gene analogs (RGAs) in Prunus: a resistance map for Prunus. Theor. Appl. Genet. 2005;111:1504–1513. doi: 10.1007/s00122-005-0079-z. [DOI] [PubMed] [Google Scholar]
- 19.Georgi LL, Wang Y, Yverggniaux D, Ormsbee T, Inigo M, Reighard GL, Abbott AG. Construction of a BAC library and its application to the identification of simple sequence repeats in peach [Prunus persica (L.) Batsch] Theor. Appl. Genet. 2002;105:1151–1158. doi: 10.1007/s00122-002-0967-4. [DOI] [PubMed] [Google Scholar]
- 20.Pampanwar V, Engler F, Hatfield J, Blundy S, Gupta G, Soderlund C. FPC Web tools for rice, maize, and distribution. Plant Physiol. 2005;138:116–126. doi: 10.1104/pp.104.056291. [DOI] [PMC free article] [PubMed] [Google Scholar]