Abstract
Better characterization of binding sites in proteins and the ability to accurately predict their location and energetic properties are major challenges which, if addressed, would have many valuable practical applications. Unfortunately, reliable benchmark datasets of binding sites in proteins are still sorely lacking. Here, we present LigASite (‘LIGand Attachment SITE’), a gold-standard dataset of binding sites in 550 proteins of known structures. LigASite consists exclusively of biologically relevant binding sites in proteins for which at least one apo- and one holo-structure are available. In defining the binding sites for each protein, information from all holo-structures is combined, considering in each case the quaternary structure defined by the PQS server. LigASite is built using simple criteria and is automatically updated as new structures become available in the PDB, thereby guaranteeing optimal data coverage over time. Both a redundant and a culled non-redundant version of the dataset is available at http://www.scmbb.ulb.ac.be/Users/benoit/LigASite. The website interface allows users to search the dataset by PDB identifiers, ligand identifiers, protein names or sequence, and to look for structural matches as defined by the CATH homologous superfamilies. The datasets can be downloaded from the website as Schema-validated XML files or comma-separated flat files.
INTRODUCTION
An increasing number of proteins with unknown function have their 3D structure solved at high resolution (1). This situation, largely due to structural genomics initiatives (2), has been stimulating the development of automated structure-based function prediction methods (3). Knowledge of residues important for function and—more particularly—for binding, can help automated prediction of function in different ways. The properties of a binding site such as its shape, atomic group or amino acid composition can provide clues on the ligand that may bind to it. Also, having information on functionally important regions in similar proteins can refine the process of annotation transfer between homologues (4).
Several methods for predicting the location of binding sites in protein structures are available and many more are currently being developed (5–7). Most of these methods need examples of known binding sites in order to derive features, which can subsequently be used to distinguish between true binding sites and sites not involved in binding. The adequate use of such methods requires the availability of valid benchmark datasets of known binding sites that can be used both to derive the appropriate combination of features and to test the prediction performance. So far, however, the availability of adequate benchmarks has been a problem.
A number of binding sites datasets have been derived from the PDB (8). But none combine enough of the necessary attributes, namely to (i) be representative of the known structural data, (ii) be non-redundant, (iii) combine information on the apo- and holo-protein structures, (iv) consist only of biologically relevant binding sites, (v) take the description of the biological unit of the protein into account, (vi) consider data from all available holo-structures for each protein and lastly (vii) be updated automatically.
Among the available datasets GOLD is a redundant set of binding sites manually checked for structural errors in the PDB files (9). SitesBase is a database of small ligand-binding sites specifically designed to enable structural comparison of known ligand-binding sites (10). The sc-PDB is a collection of binding sites specifically selected for their pharmacological interest (11). The PDBSite database attempts to annotate functional sites derived from PDB SITE records and contact residues in hetero-complexes (12). More recently, FireDB (13) has been set-up to integrate data on binding sites from the PDB and catalytic residues from the CSA (14), and organizes these data so as to allow users to assess binding site similarity in homologous proteins. To date, the only dataset of ligand-binding sites explicitly restricted to proteins with known apo-structures is the LIG dataset of 154 unique protein sequences that was used by Najmanovich et al. (15). to analyse side-chain flexibility upon ligand-binding However, the LIG dataset has—to our knowledge—not been maintained or updated, and biologically irrelevant molecules are not consistently excluded from it (with the exception of sulphate and phosphate ions).
Here, we present LigASite (‘LIGand Attachment SITE’), a dataset of ligand-binding sites in protein 3D structures, which combines all of the seven attributes listed above. In particular, it contains the description of biologically relevant binding sites in proteins with at least one apo- and one holo-structure. Information on the apo-structure is key, since due to ligand-induced conformational changes, the structure of a protein without and with the ligand (apo- and holo-structure, respectively) can differ (16,17). To mimic cases, where binding sites are truly unknown, binding site prediction methods should be validated by performing the predictions on apo-structures, and comparing them with known binding sites defined from all corresponding holo-structures. Another common problem when automatically defining binding sites from PDB holo-structures, is that some small molecules, which appear in these entries may have been introduced as part of the purification and or crystallization procedure and hence bind to the protein in a non-specific manner. Although the corresponding binding sites are usually not biologically relevant, some of these compounds may act as inhibitors or substrate analogues and hence bind to biologically relevant sites in the protein. With the aim of identifying all biologically relevant binding sites, we define them as sites bound by the biologically active ligands identified in vivo or in vitro, as well as by any compound that may act as inhibitor or substrate analogue. It is admittedly not straightforward to distinguish between such relevant ligands and non-specific binders. We however show that protocols developed in LigASite enable to automatically filter out non-specific binders with a high degree of accuracy as determined by manual validation (see Content and methods section). In all cases, these protocols take into account the quaternary structures suggested by the PQS server (18). Using the correct quaternary structures has an impact on the definition of binding sites, as these can be located at the interface between protein subunits. Optimal coverage and adequate representation of the structural data available in the PDB at any given time is enabled through fully automated procedures, and no post-processing by the user is required.
LigASite is freely accessible at http://www.scmbb.ulb.ac.be/Users/benoit/LigASite, and the data therein can be downloaded as a set of Namespace-qualified Schema-validated XML files.
CONTENT AND METHODS
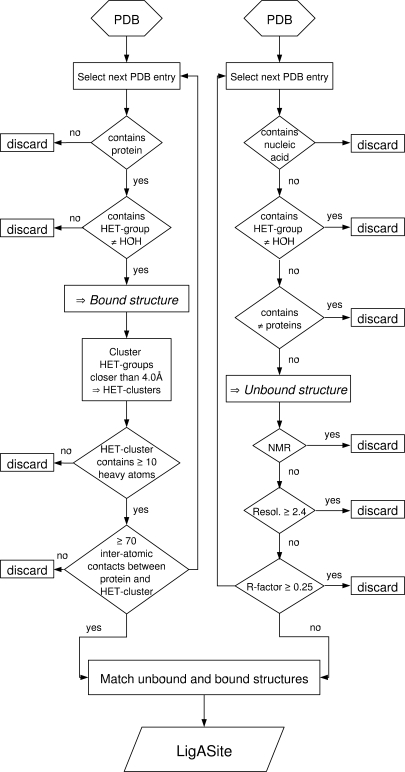
The automated procedure used to generate the LigASite dataset from the PDB is summarized in the flowchart of Figure 1.
Figure 1.
Flow-chart summarizing the automated procedure to generate LigASite from the PDB.
Generalities
Binding sites in the LigASite dataset are exclusively defined on the basis of structural data from the PDB (8). In constructing the dataset, the first step involves selecting all structures with at least one protein chain, using the PDB search engine.
As a starting point, ligands are defined as all non-water molecules that appear in the HETATM records of PDB entries. Such ligands are commonly referred to as ‘HET-groups’. Binding sites for nucleic acids and other proteins are hence ignored at this point. Quaternary structures suggested by PQS (18) are used for all structures in the dataset, unless PQS cannot be applied (e.g. NMR structures). There are 27 913 PDB entries with at least one protein chain and at least one HET-group in the PDB release of July 30, 2007, on which the current version of LigASite is based. The total number of HET-groups in these PDB entries amounts to 197 860.
Selection of biologically relevant binding sites
We define a binding site by the type of ligand that binds to it. Biologically relevant binding sites are defined as those bound by the biologically active ligands identified in vivo or in vitro, as well as by any compound that may act as inhibitor or substrate analogue. Biologically irrelevant sites are defined as sites bound non-specifically by compounds introduced by the purification or crystallization procedure.
In order to select biologically relevant sites, we filter out ligands likely to be bound non-specifically. This is done using a three-step procedure. First, HET-groups are clustered together into HET-clusters when any of their respective heavy atoms are located within <4.0 Å from one another. This clustering is performed to account for the fact that small solvent molecules often cluster together in biologically relevant binding sites and mimic parts of the natural ligand (e.g. phosphate or sulphate ions, which mimic the phosphate groups of nucleotides). Individual HET-clusters are treated here as single molecules. The total of 197 860 HET-groups in the PDB sums up to 148 879 HET-clusters.
Second, HET-clusters with less than 10 heavy atoms are rejected on the basis that biologically irrelevant molecules in PDB files are generally very small. Since HET-clusters are considered rather than single HET-groups, HET-groups with less than 10 heavy atoms, such as phosphate ions, can remain in the selected subset provided they are part of a HET-cluster with a total of at least 10 heavy atoms (e.g. two phosphate ions close to one another are kept). Overall, 52 246 HET-clusters contain at least 10 heavy atoms.
Finally, biologically relevant HET-clusters are selected based on the number of inter-atomic protein–ligand contacts. The underlying assumption being, that in general, biologically relevant ligands interact specifically with the proteins and should therefore make more inter-atomic contacts with protein atoms than their irrelevant counterparts. We use the program LPC (19) to compute the inter-atomic contacts for the following reasons: it is not restricted to a specific subset of bonds (e.g. hydrogen bonds); it provides a detailed description of contacts based on the physico-chemical properties of contacting atoms and it is freely accessible and easy to run on a large number of structures. To perform the selection based on the above assumption, we define a threshold number of inter-atomic contacts above, which an interaction is guaranteed to be relevant. To that end, the number of inter-atomic contacts between HET-cluster and protein is calculated for all HET-clusters of the dataset. The dataset is then divided into subsets based on ranges of 50 inter-atomic contacts, as indicated in Table 1. For each range, 1% of the HET-cluster–protein binding sites have been manually inspected to assess whether the binding site is biologically relevant (this manual inspection was completed for an initial version of LigASite, which was based on the December 22, 2006 PDB release). The 4th column in Table 1 shows the fractions of binding sites that we manually annotated as biologically relevant. As expected, this fraction increases as the number of inter-atomic contacts increases. Since the fraction of biologically relevant binding sites steeply increases between the ranges of 1–50 and 51–100 contacts, the subset of binding sites having between 30 and 80 contacts was subdivided into smaller bins of 10 inter-atomic contacts each, and re-analysed. Results (Table 1) show that above 70 inter-atomic contacts between HET-cluster and protein, the HET-cluster is biologically relevant in at least 95% of cases. The small number of cases where we manually annotated the binding site as biologically irrelevant even though it consists of at least 70 inter-atomic contacts between protein and HET-cluster, all correspond to membrane proteins in complex with membrane lipids, which we were therefore able to readily exclude on the basis of the ligand and/or protein type.
Table 1.
Fractions of biologically relevant binding sites as a function of the number of inter-atomic contacts between HET-cluster and protein, for steps of 50 inter-atomic contacts and steps of 10 inter-atomic contacts
| No. of contactsa | No. of sitesb | No. of inspectedc | Frelevantd |
|---|---|---|---|
| Steps of 50 inter-atomic contacts | |||
| 1–50 | 6825 | 68 | 0.54 |
| 51–100 | 15326 | 153 | 0.93 |
| 101–150 | 9786 | 98 | 0.94 |
| 151–200 | 6531 | 65 | 0.98 |
| >200 | 3932 | 39 | 0.98 |
| Steps of 10 inter-atomic contacts | |||
| 31–40 | 2145 | 21 | 0.10 |
| 41–50 | 2793 | 28 | 0.32 |
| 51–60 | 3287 | 33 | 0.64 |
| 61–70 | 3129 | 31 | 0.77 |
| 71–80 | 3293 | 33 | 0.97 |
The manual analysis summarised in this table was conducted using an initial version of LigASite, which was based on the PDB release of December 22, 2006.
aRange of number of inter-atomic contacts between protein and HET-cluster.
bTotal number of binding sites where number of inter-atomic contacts between protein and HET-cluster is within range given in column 1.
cNumber of binding sites inspected manually from the literature, used for assessing biological relevance of HET-cluster. This number equals 1% of the number of sites in column 2.
dFraction of manually inspected binding sites that we annotated as biologically relevant.
The resulting dataset is thus limited to binding sites for HET-clusters that consist of at least 10 heavy atoms, and that make at least 70 inter-atomic contacts with protein atoms. There are 32 656 such HET-clusters, consisting of 63 105 individual HET-groups and distributed among 14 459 PDB entries.
Selection of proteins with both apo- and holo-structures
A gold-standard dataset of binding sites should include information on both the apo- and holo-forms of the protein, to enable taking into account the structural changes that can occur in a protein upon binding of its ligand(s).
In order to derive the list of ‘true’ apo-structures in the PDB, we applied the following filtering procedure. From the PDB release of July 30, 2007, we selected all PDB entries with at least one protein chain that is not in complex with any other molecule, the latter being a small molecule, another protein or a nucleic acid chain. This procedure resulted in a set of 10 046 PDB entries, which was then filtered to remove Cα-only and non-X-ray entries and X-ray entries with resolution better or equal to 2.4 Å, and R-value better or equal to 0.25. These quality filters were imposed to ensure that the apo-structures, which should be used to apply functional site prediction methods, are of sufficient quality to serve as input for any type of methods including those that are based on energy calculations (20). The final set of high quality apo-structures consist of 3995 PDB entries.
These apo-structures are then paired with the corresponding holo-structures containing the biologically relevant binding sites when they share 100% sequence identity, thus resulting in a set of 550 entries with one apo-structure and at least one holo-structure with a biologically relevant binding site. Removing redundancy at 25% sequence identity [using PISCES (21)] results in a non-redundant set of 286 proteins.
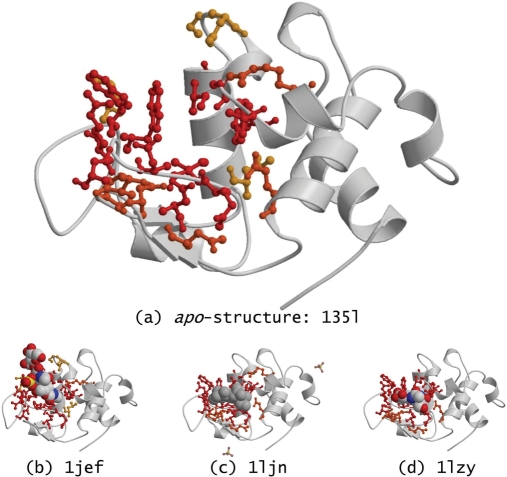
When several holo-structures are available for a given protein, all are used to define its binding site. Many holo-structures are bound to only a portion of the natural ligands, and the picture of the binding site that is obtained when considering all available holo-structures is therefore more complete and accurate. Out of the 550 proteins in the redundant dataset, 291 have more than one holo-structure. A frequency score is assigned to all binding site residues in a protein, based on the fraction of corresponding holo-structures in which the residue is observed to be part of a biologically relevant binding site (Figure 2).
Figure 2.
Turkey egg-white lysozyme. (a) Mapping of binding site residues on the apo-structure (PDB entry 135l). These residues are identified as part of the binding site from the three holo-structures in which the lysozyme is in complex with different ligands: (b) with three molecules of N-acetyl-d-glucosamine (NAG) and a sulphate ion in PDB entry 1jef; (c) with di(N-acetyl-d-glucosamine) in PDB entry 1ljn; and (d) with two molecules of NAG in PDB entry 1lzy. Binding site residues are coloured red if they are in contact with ligand atoms in all three holo-structures (i.e. frequency score of 1.0 = 3/3), they are coloured orange if they are only in contact with ligand atoms in two of the holo-structures (i.e. frequency score of 0.67 ≈ 2/3), and they are coloured yellow if they are in contact with ligand atoms in only one holo-structure (i.e. frequency score of 0.33 ≈ 1/3). HET-groups considered as biologically relevant in LigASite are displayed and coloured in CPK. Sulphate ions filtered out as biologically irrelevant in PDB entry 1ljn are transparent and displayed in balls-and-sticks. The figure was drawn with molscript (27) and rendered with Raster3D (28).
Website implementation
The fact that the construction of the dataset only relies on simple numerical cut-offs and automatic filters allows it to be automatically updated as new data become available in the PDB. At the time of writing the manuscript, the current release of LigASite is based on the July 30, 2007 release of the PDB. LigASite is updated monthly.
LigASite is accessible at the following URL: http://www.scmbb.ulb.ac.be/Users/benoit/LigASite. Both the redundant and the non-redundant versions of the dataset can be browsed. For each protein, a front-page describes the apo-structure, and lists all binding site residues defined from the ensemble of corresponding holo-structures. Binding site residue positions (i.e. PDB serial numbers) are coloured on a yellow-to-red scale, depending on the fraction of holo-structures in which the residue is found in contact with a biologically relevant HET-cluster (red when the fraction equals 1). Residue three-letter codes are coloured according to their physico-chemical type. PDB identifiers of corresponding holo-structures are listed together with unique identifiers describing the biologically relevant HET-clusters used to define the binding site residues, the number of heavy atoms in the HET-clusters and the number of inter-atomic contacts (identified with LPC) between each HET-cluster and protein atoms. A PDB file providing coordinates of the binding site residues in the apo-structure can be downloaded for each protein.
Clicking on the ‘Details’ links associated with holo-structures leads to a holo-structure specific page, on which more details can be found for each structure. Residues identified as part of binding sites in the given holo-structure are listed, together with the unique identifier of the HET-group with which each residue interacts. Residues interacting with HET-groups in other holo-structures of the same protein are not listed. Further data on the HET-groups that bind in biologically relevant binding sites is also available from these pages, i.e. PDB HET identifiers, molecule name, chemical formula and (non-stereo) SMILES string. A PDB file providing coordinates of the binding site residues and of the HET-groups labelled as biologically relevant is available for download for each holo-structure in the dataset.
Cross-links to relevant databases [e.g. PDBsum (22), CSA (14)] are available on all pages, as well as links to LIGPLOT (23) drawings of the interactions between HET-groups and proteins. A search facility is available and allows users to search LigASite for PDB identifiers (both apo- and holo-), HET-group identifiers (e.g. ‘ATP’) or protein names. The LigASite dataset can also be searched by sequence similarity using BLAST (24). All matches to a query sequence are returned to the user and are sorted by sequence identity and E-value. Finally, LigASite can be searched for proteins that have a domain in the same CATH superfamily (i.e. H level) as the domains in the PDB entry input by the user (25).
XML is a markup language that combines text and information about this text. XML Schema allows to define semantic constraints for the data contained in the XML files. An XML file is not only humanly-readable, but can also be easily parsed, and consequently validated and transformed, by a computer. An XML Schema (‘LigASiteML’) has been designed for a complete description of binding site information on the proteins of the dataset. An XML file describing the complete LigASite database is generated for each update, and validated against the above-mentioned XML Schema. Both the Schema and XML files are available for download from the website, allowing users to easily use LigASite data locally. In addition, protein-specific XML files, validated against the same XML Schema, and describing only binding site residues in a particular protein of the dataset are available for download from the pages describing the apo- and holo-structures of the corresponding protein. A comma-separated file containing the core data of LigASite, and created automatically from the XML file, is also available for download.
Some Statistics on dataset content
The HET-groups featured in the biologically relevant binding sites of LigASite proteins are very diverse. In total, 551 different HET-groups appear in the holo-structures of the non-redundant LigASite dataset (redundancy removed at 25% sequence identity). This number exceeds the number of binding sites in the dataset because (i) several different HET-groups can appear together in a binding site in a single holo-structure, e.g. a Magnesium ion (HET ID ‘MG’) and a molecule of adenosine-5′-diphosphate (HET ID ‘ADP’) in the holo-structure 1pfk of phosphofructokinase; and (ii) different HET-groups can appear in a given binding site in different holo-structures of a protein (Figure 2). The majority of HET-groups appear in the binding site of only one protein, suggesting an important diversity of ligands in the dataset.
Table 2 gives the names, HET IDs and numbers of occurrences of the HET-groups that appear most frequently in binding sites of the non-redundant version of LigASite. When a HET-group appears in several holo-structures of a given protein, only one occurrence is counted. Therefore, the ‘number of occurrences’ in this table corresponds to the number of different proteins to which a given HET-group binds. The most common HET-group is the Magnesium ion, which occurs in the binding sites of 41 different proteins, as part of HET-clusters consisting of more than 10 atoms (e.g. ‘MG’ together with ‘ATP’ constitute the HET-cluster in the binding site of PDB entry 1e4g). Several nucleotides and derivatives thereof are also among the most frequent HET-groups in LigASite (i.e. ATP, ADP, AMP, GDP, COA, NAD and NAP). Interestingly, a number of small molecules commonly ignored in existing datasets of binding sites because of their potential irrelevance, appear in several binding sites in LigASite (i.e. phosphate ions and glycerol molecules, and sulphate ions which appear in the binding sites of six different proteins).
Table 2.
Most frequent HET-groups in LigASite (nr25)
| Namea | HET IDb | No. of occurrencesc |
|---|---|---|
| Magnesium ion | MG | 41 |
| Adenosine-5′-diphosphate | ADP | 19 |
| Manganese (II) ion | MN | 13 |
| Adenosine-5′-triphosphate | ATP | 13 |
| N-Acetyl-d-glucosamine | NAG | 12 |
| Nicotinamide-adenine-dinucleotide | NAD | 11 |
| Glycerol | GOL | 11 |
| Guanosine-5′-diphosphate | GDP | 11 |
| Phosphate ion | PO4 | 9 |
| Glucose | GLC | 9 |
| NADP | NAP | 8 |
| d-Galactose | GAL | 8 |
| Coenzyme A | COA | 8 |
| Adenosine monophosphate | AMP | 8 |
| Phosphoaminophosphonic acid-adenylate ester | ANP | 7 |
aHET-group name as provided in the PDB Chemical Component Dictionary (see http://deposit.pdb.org/cc_dict_tut.html).
bHET-group ID (i.e. ‘residue name’ in PDB files).
cNumber of occurrences of HET-groups in LigASite (redundancy removed at 25% seq. id.). When a given HET-group appears in several different holo-structures of a given protein, only one occurrence was counted to compute values in this table.
We used the PDBSProtEC mapping (26), in order to obtain the EC numbers for all 286 proteins in the non-redundant version of LigASite, and for all the proteins of a non-redundant version of the PDB [redundancy removed at 25% sequence identity using PISCES (21)] (Figure 3). Only 48 proteins out of 286 (i.e. 17%) in the non-redundant version of LigASite are non-enzymes (26). In the non-redundant version of the PDB, 39% of proteins are non-enzymes. Among enzymes, transferases and hydrolases (EC classes 2. and 3., respectively) are the most common both in LigASite and the PDB.
Figure 3.
Distribution of EC classes among proteins in (a) the non-redundant version of the LigASite dataset (redundancy removed at 25% sequence identity), and in (b) a non-redundant subset of the PDB (redundancy removed at 25% sequence identity), which consists of 5180 PDB entries. EC numbers were obtained from PDBSprotEC, a mapping of PDB entries to EC numbers via SwissProt (26).
CONCLUSIONS
LigASite is a publicly available dataset of binding sites in proteins with at least one known apo-structure and one known holo-structure. Biologically irrelevant binding sites are automatically filtered out from the dataset with high accuracy. The dataset relies on simple numerical cut-offs and is automatically updated regularly.
In its current version, LigASite contains binding sites for all ligands stored as HET-groups in the PDB (e.g. small organic compounds, oligo-saccharides, lipids, nucleotides and derivatives thereof, etc.). We are presently working on extending our protocol, in order to include binding sites for peptides, proteins and nucleic acids. We furthermore plan to derive an improved score for excluding biologically irrelevant binding sites from the dataset. This score, in which the number of inter-atomic contacts between protein and ligand (i.e. the one currently used) is normalized by the number of heavy atoms in the ligand, should allow us to increase our coverage of the small biologically relevant ligands, which make few contacts with protein atoms (Table 1). We are also working on the development of a structure-based search facility to allow users to look for local structural matches with binding sites in LigASite.
LigASite should prove a highly valuable resource for validating and developing binding site prediction methods, and for the study of binding site properties in general.
ACKNOWLEDGEMENT
The EU 6th Framework Program is gratefully acknowledged for support, through the GeneFun ‘In-silico prediction of gene function’ and BioSapiens Network of Excellence projects, under the thematic area ‘Life sciences, genomics and biotechnology for health’, contract numbers: LSHG-CT-2004-503567 and LSHG-CT-2003-503265. Funding to pay the Open Access publication charges was provided by the EU 6th Framework Programme through the GeneFun (LSHG-CT-2004-503567) project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rigden DJ. Understanding the cell in terms of structure and function: insights from structural genomics. Curr. Opin. Biotechnol. 2006;17:457–464. doi: 10.1016/j.copbio.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Skolnick J, Fetrow JS, Kolinski A. Structural genomics and its importance for gene function analysis. Nat. Biotechnol. 2000;18:283–287. doi: 10.1038/73723. [DOI] [PubMed] [Google Scholar]
- 3.Watson JD, Laskowski RA, Thornton JM. Predicting protein function from sequence and structural data. Curr. Opin. Struct. Biol. 2005;15:275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Polacco BJ, Babbitt PC. Automated discovery of 3D motifs for protein function annotation. Bioinformatics. 2006;22:723–730. doi: 10.1093/bioinformatics/btk038. [DOI] [PubMed] [Google Scholar]
- 5.Jones S, Thornton JM. Searching for functional sites in protein structures. Curr. Opin. Chem. Biol. 2004;8:3–7. doi: 10.1016/j.cbpa.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Laurie AT, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 7.Glaser F, Morris RJ, Najmanovich RJ, Laskowski RA, Thornton JM. A method for localizing ligand binding pockets in protein structures. Proteins. 2006;62:479–488. doi: 10.1002/prot.20769. [DOI] [PubMed] [Google Scholar]
- 8.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nissink JW, Murray C, Hartshorn M, Verdonk ML, Cole JC, Taylor R. A new test set for validating predictions of protein-ligand interaction. Proteins. 2002;49:457–471. doi: 10.1002/prot.10232. [DOI] [PubMed] [Google Scholar]
- 10.Gold ND, Jackson RM. SitesBase: a database for structure-based protein-ligand binding site comparisons. Nucleic Acids Res. 2006;34:D231–234. doi: 10.1093/nar/gkj062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kellenberger E, Muller P, Schalon C, Bret G, Foata N, Rognan D. sc-PDB: an annotated database of druggable binding sites from the Protein Data Bank. J. Chem. Inf. Model. 2006;46:717–727. doi: 10.1021/ci050372x. [DOI] [PubMed] [Google Scholar]
- 12.Ivanisenko VA, Pintus SS, Grigorovich DA, Kolchanov NA. PDBSite: a database of the 3D structure of protein functional sites. Nucleic Acids Res. 2005;33:D183–187. doi: 10.1093/nar/gki105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez G, Valencia A, Tress M. FireDB–a database of functionally important residues from proteins of known structure. Nucleic Acids Res. 2007;35:D219–223. doi: 10.1093/nar/gkl897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porter CT, Bartlett GJ, Thornton JM. The catalytic site atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Najmanovich R, Kuttner J, Sobolev V, Edelman M. Side-chain flexibility in proteins upon ligand binding. Proteins. 2000;39:261–268. doi: 10.1002/(sici)1097-0134(20000515)39:3<261::aid-prot90>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Muller CW, Schlauderer GJ, Reinstein J, Schulz GE. Adenylate kinase motions during catalysis: an energetic counterweight balancing substrate binding. Structure. 1996;4:147–156. doi: 10.1016/s0969-2126(96)00018-4. [DOI] [PubMed] [Google Scholar]
- 17.Magnusson U, Chaudhuri BN, Ko J, Park C, Jones TA, Mowbray SL. Hinge-bending motion of D-allose-binding protein from Escherichia coli: three open conformations. J. Biol. Chem. 2002;277:14077–14084. doi: 10.1074/jbc.M200514200. [DOI] [PubMed] [Google Scholar]
- 18.Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem. Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 19.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 20.Dessailly BH, Lensink MF, Wodak SJ. Relating destabilizing regions to known functional sites in proteins. BMC Bioinformatics. 2007;8:141. doi: 10.1186/1471-2105-8-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, Dunbrack R.L., Jr. PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005;33:D266–268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin AC. PDBSprotEC: a web-accessible database linking PDB chains to EC numbers via SwissProt. Bioinformatics. 2004;20:986–988. doi: 10.1093/bioinformatics/bth048. [DOI] [PubMed] [Google Scholar]
- 27.Kraulis PJ. Molscript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 28.Merritt EA, Bacon DJ. Raster3D version 2: photorealistic molecular graphics. Meth. Enz. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]