Abstract
The starlet sea anemone, Nematostella vectensis, is a basal metazoan organism that has recently emerged as an important model system in developmental biology and evolutionary genomics. StellaBase, the Nematostella Genomics Database (http://stellabase.org), was developed in 2005 as a resource to support the Nematostella research community. Recently, it has become apparent that Nematostella may be a particularly useful system for studying (i) microevolutionary variation in natural populations, and (ii) the functional evolution of human disease genes. We have developed two new databases that will foster such studies: StellaBase Disease (http://stellabase.org/disease) is a relational database that houses 155 904 invertebrate homologous isoforms of human disease genes from four leading genomic model systems (fly, worm, yeast and Nematostella), including 14 874 predicted genes from the sea anemone itself. StellaBase SNP (http://stellabase.org/SNP) is a relational database that describes the location and underlying type of mutation for 20 063 single nucleotide polymorphisms.
INTRODUCTION
The starlet sea anemone, Nematostella vectensis, is a member of the basal metazoan phylum Cnidaria. This species is emerging as an important model system in evolutionary genomics, developmental biology and estuarine ecology due to (i) the availability of a complete genome sequence, (ii) the generally conservative evolution of its genome, (iii) its ease of culture, (iv) the experimental tractability of its entire life history and (v) the ease of collecting the animal from the field. The increasing utility of this species is evident from the recent surge in research papers—a PubMed search using the query ‘nematostella’ returns 35 journal articles published in 2006–07.
StellaBase is a genomic database for Nematostella that allows users to search the genome sequence, predicted genes, predicted proteins cross-referenced with PFAM motifs (1) and expressed sequence tags (ESTs) (2). The database is built on a relational structure in MySQL with a front-end HTML interface on an Apache server. It also houses a primer database, a literature database and a database of living genetic stocks and DNA samples that is searchable by geographic locale or by population-specific genetic markers. Many recent publications on Nematostella have been facilitated by this relational database.
The utility of a sequenced genome depends partially on the rate at which it has evolved. Slowly evolving taxa are more informative about ancient evolutionary events, whereas rapidly evolving taxa can prove useful for microevolutionary studies. The Nematostella genome is proving to be useful for reconstructing both ancient and recent evolutionary events because the genome appears to have evolved relatively slowly on a macroevolutionary scale, but relatively rapidly on a microevolutionary scale. For example, Nematostella shares far more intron locations with humans than do Drosophila melanogaster, Anopheles gambiae or Caenorhabditis elegans (3,4). It also shares more orthologous genes with humans than these protostome animals and even the sea squirt Ciona intestinalis, which, like human, is a chordate (3). At the same time, Nematostella exhibits a relatively high rate of intraspecific polymorphism and extensive population structure at very fine spatial scales (3,5).
STELLABASE DISEASE
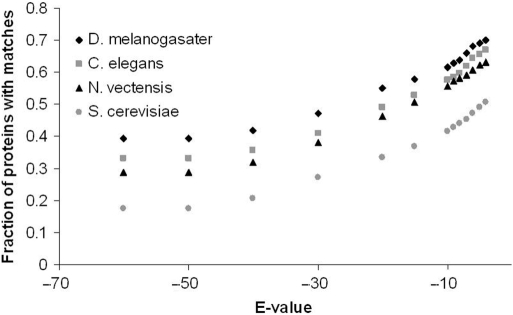
The remarkable degree of genomic conservation between the starlet anemone and humans suggests that Nematostella could become a useful model system for exploring the functional evolution of proteins underlying human disease. Nematostella's genome contains about the same number of homologs to human disease genes as do D. melanogaster and C. elegans [Figure 1; (6)], despite the fact that the fruitfly and soil nematode are more closely related to humans by 50 million years or more. Furthermore, in several instances where all three of these invertebrate model systems possess an ortholog to the same disease-causing gene in humans, the Nematostella variant can be far more similar to the human form. For example over the 1168 amino acid region spanning its eight breast cancer repeats, the human breast cancer related gene BRCA2 is 15% identical and 29% similar to its Nematostella homolog but only 6% identical and 13% similar to its D. melanogaster homolog (6).
Figure 1.
The fraction of human disease genes that have at least one putative homolog in anemone, fruitfly, nematode worm and yeast. Putative homologs were identified using BLASTP searches across a range of E-value thresholds (depicted along the X-axis). Although the log-scale represents E-values only as small as 1e-60, no change is seen in the resulting number of hits for any taxa when the E-value threshold is varied between 1e-60 and 1e-100.
Many invertebrate models offer considerable advantages over vertebrates for the performance of systems level research, and for this reason, they have proven useful for understanding the function of proteins underlying human disease states. Fruitfly and soil nematode, for example, are far less expensive to culture than vertebrates, they can be raised in far greater numbers, and their generation times are shorter. Their greater experimental tractability offsets their greater phylogenetic distance from humans. Nematostella shares many of these same experimental advantages (5), and given its surprisingly high degree of genomic similarity to human (3,4,6), Nematostella may prove particularly useful for illuminating the functional evolution of disease genes (6). Furthermore, Nematostella is capable of complete bi-directional regeneration (7), which allows the rapid production of clonal genetic stocks in the laboratory, a pronounced experimental advantage not shared by the fruitfly or the soil nematode.
That we might better exploit the starlet sea anemone for its potential to inform the molecular understanding of human diseases, we identified orthologs of human disease genes from the sequenced genomes of N. vectensis, D. melanogaster, C. elegans and Saccharomyces cerevisiae, and we compiled them in a relational database integrated with StellaBase. Comparative genomic databases have been developed for this purpose in the past [e.g. (8,9)] but none of these include data from N. vectensis, or any other basal metazoans.
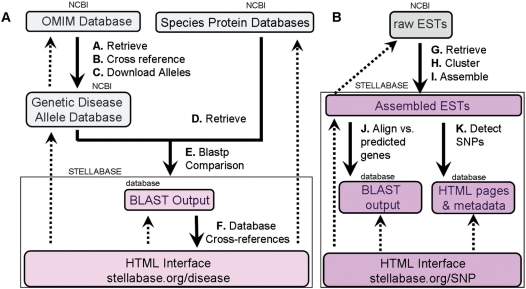
To develop StellaBase Disease, we downloaded the OMIM database and select tables from GenBank (Genbank tables: gene2accession and mim2gene; Figure 2a, Step ‘A’). Cross-referencing between genemap, mim2gene and gene2accession allowed for the identification and downloading of all alleles associated with the 10 237 OMIM entries available on 15 July 2007 (Figure 2a, Steps ‘B’ and ‘C’). The protein datasets for fly, nematode and baker's yeast were downloaded from NCBI (Figure 2a, Step ‘D’). These datasets, and the predicted proteins available in StellaBase v.1.0, were formatted as blast databases and were searched with the retrieved human disease allele database using BLASTP (10). The resultant BLASTP output and OMIM data are stored in a relational database cross-referenced with the main structure of StellaBase (Figure 3).
Figure 2.
Pipeline for the development of (A) StellaBase Disease and (B) StellaBase SNP.
Figure 3.
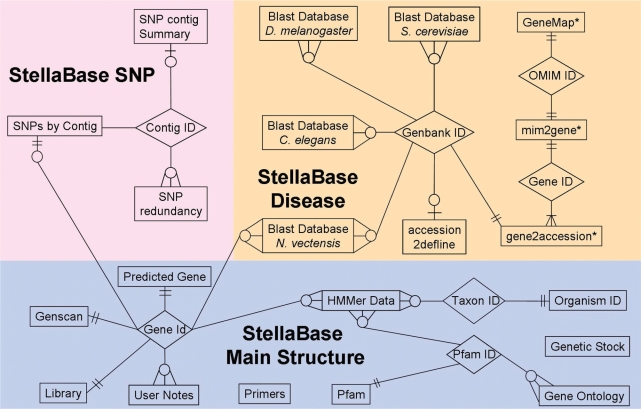
Entity-relationship diagram for StellaBase. Tables are represented by rectangles, and interfaces among tables are represented by diamonds. Tables marked with an asterisk have been downloaded from NCBI. (cardinality: 2 straight lines, exactly 1; circle, zero; crow's feet, more than 1; circle and line, 0 or 1; circle and crow's feet, 0 or many; line and crow's feet, 1 or many).
These databases are searchable via an html interface (http://stellabase.org/disease) using OMIM identification numbers, GenBank identification numbers and StellaBase protein identification numbers. Keyword searches allow users to browse any OMIM entry with the search term included within the OMIM description. Results can be filtered by taxon and significance scores for blast hits. Organism-specific summary tables allow users to retrieve specific results for individual organisms and individual alleles, and to obtain additional data for each locus that displays a significant match to any human disease allele query.
The 10 237 OMIM entries referenced in StellaBase Disease refer to 142 500 human disease alleles. A large fraction of these genes are predicted to have orthologs in the model invertebrate taxa indexed in StellaBase Disease (Figure 1). The database houses homology data for 155 904 potential invertebrate allelic orthologs. It is significant that 3670 of the human disease allelic variants lack a homolog in fly, worm and yeast yet possess a putative homolog in Nematostella (at an E-value of 1e-4). For researchers studying these proteins, which corresponds to ∼2.58% of the OMIM database, Nematostella may be the only established model invertebrate system available.
STELLABASE SNP
The abundant intraspecific polymorphism harbored by Nematostella likely results from a combination of three factors—a wide geographic distribution aided by anthropogenic dispersal, low levels of natural dispersal between estuaries and even within estuaries (11) and local adaptation to diverse environments. We can therefore utilize the genomic variation present in this species to address a range of important ecological and evolutionary questions including the natural and human-aided dispersal of coastal invertebrates, the evolution of native versus introduced populations and the microevolutionary basis for organismal adaptations to key environmental variables including temperature, salinity and pH. Indeed, the cells of Nematostella must encounter a greater range of natural variation in these key variables than any other animal genomic model system, making Nematostella a uniquely informative model system in which to study adaptation to these variables.
To facilitate the study of intraspecific genetic diversity in Nematostella and allow researchers to identify functionally significant polymorphisms, we have developed a polymorphism database for Nematostella expressed sequences. The polymorphisms were identified in 16 619 ESTs downloaded from NCBI on 1 February 2007 (Figure 2b, Step ‘G’). These ESTs were clustered and assembled using the TIGR Gene Indices clustering tool (12). A total of 20 063 potential SNPs were identified in 3233 EST builds and indexed using AutoSNP (13,14). All of the SNPs are stored in a relational database cross-referenced with the main structure of StellaBase (Figure 3).
Data housed in StellaBase SNP can be accessed through (i) a blast search, (ii) a browse function and (iii) an html query page. (i) The blast search page of StellaBase SNP (http://stellabase.org/SNP/blast/blast_cs.html) allows users to search both the raw ESTs and the 3233 assembled ESTs (contigs). Data associated with each contig can then be retrieved either through the (ii) ‘browse contig function’ (http://stellabase.org/SNP/autoSNP/contigsummary.html) or (iii) the HTML query page (http://stellabase.org/SNP). We cross-referenced each EST contig with the corresponding predicted gene in StellaBase using BLASTN (Figure 2b, Step ‘J’). The cross-referencing allows users to search the ESTs using a StellaBase ID or an associated Pfam motif. Because the ESTs do not commonly span the entire protein-coding region, it might not be possible to associate them directly with particular Pfam motifs. Cross-referencing them with the predicted genes in StellaBase can overcome this limitation in some instances. Users are able to browse and search for appropriate Pfam motifs based upon keyword searches to identify the correct Pfam accession number or Pfam name to use in a query.
HTML queries can be filtered based upon three criteria. First, the query can return only those assembled ESTs that match a predicted protein housed in StellaBase (and/or a given Pfam motif) at a threshold E-value. This search modality is disabled unless users are querying for a particular protein motif. Second, the query can return only those polymorphic ESTs exhibiting a minimum SNP redundancy. The minimum redundancy filter limits output to only those SNPs for which the less abundant variant has been identified in ‘at least’ the number of ESTs specified by the user. Third, users can limit the results to include only those assembled ESTs that contain a user-defined minimum number of polymorphic sites. Both ‘minimum SNP redundancy’ and ‘minimum number of polymorphic sites’ can be employed as filters, allowing users to browse all assembled ESTs that meet or exceed threshold values.
Assembled ESTs can be retrieved through a download tool (http://stellabase.org/SNP/get_est_contig.cgi). Additional information about each locus can be retrieved through linked StellaBase matches or the StellaBase gene search page (http://stellabase.org/html/gene_search.html) once a corresponding genomic locus is identified.
GENE ONTOLOGY CROSS-REFERENCES
In addition to the disease gene and SNP databases, we have added Gene Ontology cross-references to StellaBase (http://stellabase.org/html/GO.html). To accomplish this, we downloaded the Gene Ontology database (15), and used SQL scripting to create a simple table to cross-reference GO terms with Pfam motifs. This table was incorporated into StellaBase (Figure 3), allowing StellaBase to be searched using GO terms.
The incorporation of Gene Ontology terms allows for much more complex queries than would be possible using Pfam motifs alone. Users can retrieve all predicted Nematostella proteins whose GO terms include broad descriptors of protein functionality or localization, i.e. ‘nucleus’. The potential complexity of these queries precludes completion of the query within an appropriate timeframe for HTML/CGI scripting. As such, results from user queries are e-mailed to users, in a user-specified data output format.
CONCLUSIONS AND FUTURE DIRECTIONS
The emergence of Nematostella vectensis as a model system began with the developmental and field studies of Hand and Uhlinger in the early 1990s (16–18), and it gained momentum with the publication of the first molecular studies (19,20), which demonstrated that the starlet sea anemone could yield key insights into early animal evolution. StellaBase was developed primarily with the evolutionary genomics community in mind. However, recent studies demonstrate that the utility of Nematostella as a model system extends to microevolutionary studies and even medical research. The upgrades described here are intended to serve these communities. We intend to augment the functionality of StellaBase and perform regular upgrades into the future, and we welcome user comments on how the database might be improved.
ACKNOWLEDGEMENTS
This work was supported by NSF grant FP-91656101-0 to J.C.S. and J.R.F. and EPA Grant F5E11155 to A.R.M. and J.R.F. and by a Postdoctoral Scholar Program at the Woods Hole Oceanographic Institution, with funding provided by The Beacon Institute for Rivers and Estuaries, and the J. Seward Johnson Fund to A.M.R. J.C.S. would like to thank J.F. Ryan for his very patient and persistent tutelage. Funding to pay the OpenAccess publication charges for this article was provided by a grant from the Marine Management Area Science Program of Conservation International.
Conflict of interest statement. None declared.
REFERENCES
- 1.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan JC, Ryan JF, Watson JA, Webb J, Mullikin JC, Rokhsar D, Finnerty JR. StellaBase: the Nematostella vectensis Genomics Database. Nucleic Acids Res. 2006;34:D495–D499. doi: 10.1093/nar/gkj020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan JC, Reitzel AM, Finnerty JR. A high percentage of introns in human genes were present early in animal evolution: evidence from the basal metazoan Nematostella vectensis. Genome Inf. 2006;17:219–229. [PubMed] [Google Scholar]
- 5.Darling JA, Reitzel AR, Burton PM, Mazza ME, Ryan JF, Sullivan JC, Finnerty JR. Rising starlet: the starlet sea anemone, Nematostella vectensis. Bioessays. 2005;27:211–221. doi: 10.1002/bies.20181. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan JC, Finnerty JR. A surprising abundance of human disease genes in a simple basal animal, the starlet sea anemone (Nematostella vectensis) Genome. 2007;50:689–692. doi: 10.1139/g07-045. [DOI] [PubMed] [Google Scholar]
- 7.Reitzel AM, Burton P, Krone C, Finnerty JR. Comparison of developmental trajectories in the starlet sea anemone Nematostella vectensis: embryogenesis, regeneration, and two forms of asexual fission. Invertebr. Biol. 2007;126:99–112. [Google Scholar]
- 8.Chien S, Reiter LT, Bier E, Gribskov M. Homophila: human disease gene cognates in Drosophila. Nucleic Acids Res. 2002;30:149–151. doi: 10.1093/nar/30.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Brien KP, Westerlund I, Sonnhammer EL. OrthoDisease: a database of human disease orthologs. Hum. Mutat. 2004;24:112–119. doi: 10.1002/humu.20068. [DOI] [PubMed] [Google Scholar]
- 10.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darling JA, Reitzel AM, Finnerty JR. Regional population structure of a widely introduced estuarine invertebrate: Nematostella vectensis Stephenson in New England. Mol. Ecol. 2004;13:2969–2981. doi: 10.1111/j.1365-294X.2004.02313.x. [DOI] [PubMed] [Google Scholar]
- 12.Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 13.Batley J, Barker G, O'Sullivan H, Edwards KJ, Edwards D. Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data. Plant Physiol. 2003;132:84–91. doi: 10.1104/pp.102.019422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savage D, Batley J, Erwin T, Logan E, Love CG, Lim GA, Mongin E, Barker G, Spangenberg GC, et al. SNPServer: a real-time SNP discovery tool. Nucleic Acids Res. 2005;33:W493–W495. doi: 10.1093/nar/gki462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hand C, Uhlinger K. The culture, sexual and asexual reproduction, and growth of the sea anemone Nematostella vectensis. Biol. Bull. 1992;182:169–176. doi: 10.2307/1542110. [DOI] [PubMed] [Google Scholar]
- 17.Hand C, Uhlinger K. The unique, widely distributed sea anemone, Nematostella vectensis Stephenson: A review, new facts, and questions. Estuaries. 1994;17:501–508. [Google Scholar]
- 18.Hand C, Uhlinger KR. Asexual reproduction by transverse fission and some anomalies in the sea anemone Nematostella vectensis. Invertebr. Biol. 1995;114:9–18. [Google Scholar]
- 19.Finnerty JR, Martindale MQ. Homeoboxes in sea anemones (Cnidaria:Anthozoa): a PCR-based survey of Nematostella vectensis and Metridium senile. Biol. Bull. 1997;193:62–76. doi: 10.2307/1542736. [DOI] [PubMed] [Google Scholar]
- 20.Finnerty JR, Martindale MQ. Ancient origins of axial patterning genes: Hox genes and ParaHox genes in the Cnidaria. Evol. Dev. 1999;1:16–23. doi: 10.1046/j.1525-142x.1999.99010.x. [DOI] [PubMed] [Google Scholar]