Abstract
The recent SARS epidemic has boosted interest in the discovery of novel human and animal coronaviruses. By July 2007, more than 3000 coronavirus sequence records, including 264 complete genomes, are available in GenBank. The number of coronavirus species with complete genomes available has increased from 9 in 2003 to 25 in 2007, of which six, including coronavirus HKU1, bat SARS coronavirus, group 1 bat coronavirus HKU2, groups 2c and 2d coronaviruses, were sequenced by our laboratory. To overcome the problems we encountered in the existing databases during comparative sequence analysis, we built a comprehensive database, CoVDB (http://covdb.microbiology.hku.hk), of annotated coronavirus genes and genomes. CoVDB provides a convenient platform for rapid and accurate batch sequence retrieval, the cornerstone and bottleneck for comparative gene or genome analysis. Sequences can be directly downloaded from the website in FASTA format. CoVDB also provides detailed annotation of all coronavirus sequences using a standardized nomenclature system, and overcomes the problems of duplicated and identical sequences in other databases. For complete genomes, a single representative sequence for each species is available for comparative analysis such as phylogenetic studies. With the annotated sequences in CoVDB, more specific blast search results can be generated for efficient downstream analysis.
INTRODUCTION
Coronaviruses are found in a wide variety of animals and are associated with respiratory, enteric, hepatic and neurological diseases of varying severity. Based on genotypic and serological characterization, coronaviruses were divided into three distinct groups (1–3). As a result of the unique mechanism of viral replication, coronaviruses have a high frequency of recombination (2,4).
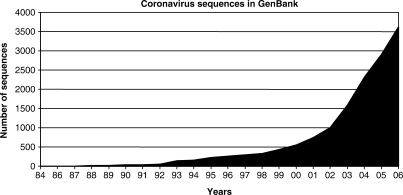
The recent severe acute respiratory syndrome (SARS) epidemic, the discovery of SARS coronavirus (SARS-CoV) and identification of SARS-CoV-like viruses from Himalayan palm civets and a raccoon dog from wild live markets in China have led to a boost in interest on discovery of novel coronaviruses in both humans and animals (5–9) (Figure 1). For human coronaviruses, a novel group 1 human coronavirus, human coronavirus NL63 (HCoV-NL63) was reported in 2004 (10,11), while we described the discovery, complete genome sequence and genetic diversity of a novel group 2 human coronavirus, coronavirus HKU1 (CoV-HKU1) in 2005 (4,12–14). As for animal coronaviruses, six group 1 (15–17), four group 2, including bat SARS-CoV and two new subgroups of group 2 coronaviruses (6,8,18,19), and 11 group 3 (20–23) coronaviruses have recently been described.
Figure 1.
Number of coronavirus sequences in GenBank from 1984 to 2006.
By July 2007, more than 3000 coronavirus sequence records, including a total of 264 complete genomes, are available in GenBank (24). Among the 25 coronavirus species with complete genome sequence available, six were sequenced by our group, including CoV-HKU1 and bat SARS-CoV (13,16,18,19). Furthermore, we defined two novel subgroups of group 2 coronavirus (18). During the process of batch sequence retrieval for comparative genome analysis of the coronavirus genomes that we sequenced, we encountered several major problems about the coronavirus sequences in GenBank as well as other coronavirus databases (Coronaviridae Bioinformatics Resource, http://athena.bioc.uvic.ca/database.php?db=coronaviridae; PATRIC http://patric.vbi.vt.edu) (25). First, in GenBank, the non-structural proteins in the polyprotein encoded by orf1ab were not annotated. Second, in all databases, for the non-structural proteins encoded by ORFs downstream to orf1ab, the annotations are often confusing because they are not annotated using a standardized system. Third, multiple accession numbers are often present for reference sequences (26). These problems often lead to confusion when sequence retrieval is performed. Fourth, coronaviruses, especially SARS-CoV, amplified from different specimens may contain the same genome or gene sequences. These sequences usually lead to redundant work when they are analyzed.
In view of these problems, we started to develop our own database for coronavirus gene and genome sequences in 2005. In this database, CoVDB, we sought to create a user-friendly platform for efficient batch sequence retrieval, which is crucial for comparative genome analysis. In this article, we describe this comprehensive database of annotated coronavirus genes and genomes, which provides a central source of information about coronaviruses. To further increase the usefulness of CoVDB, commonly used bioinformatics tools were also included for analysis of the sequence data.
MATERIALS AND METHODS
Database description
Sequence data
CoVDB is a web-based coronavirus database. Data of CoVDB is stored and managed by MySQL database management system. By July 2007, CoVDB contains 3982 coronavirus sequences and one torovirus genome sequence. Two hundred and sixty-four of them are complete genomes and the rest are partial genomes or genes. All data were retrieved from GenBank using modules of bioperl. We annotated sequences without gene information or non-structural protein boundary and labeled the 5′ and 3′ untranslated regions (UTRs) of the genomes. By July 2007, CoVDB contains 12 344 genes and UTRs.
Information on coronavirus genome characteristics
In addition to the two sequence retrieval pages, CoVDB collects information on coronavirus sequence characteristics, including genome organization, a brief description on each complete coronavirus genome, GC content, polyprotein cleavage sites, transcription regulatory sequences, acidic tandem repeat sequences and known RNA structures. These pieces of information can be accessed by clicking ‘Genome’ in the top menu bar of CoVDB. In the ‘Tools’ page, blast similarity search (27) against annotated coronavirus sequences in CoVDB can be performed and other commonly used tools are also provided.
Functionality of the database
Batch sequence retrieval
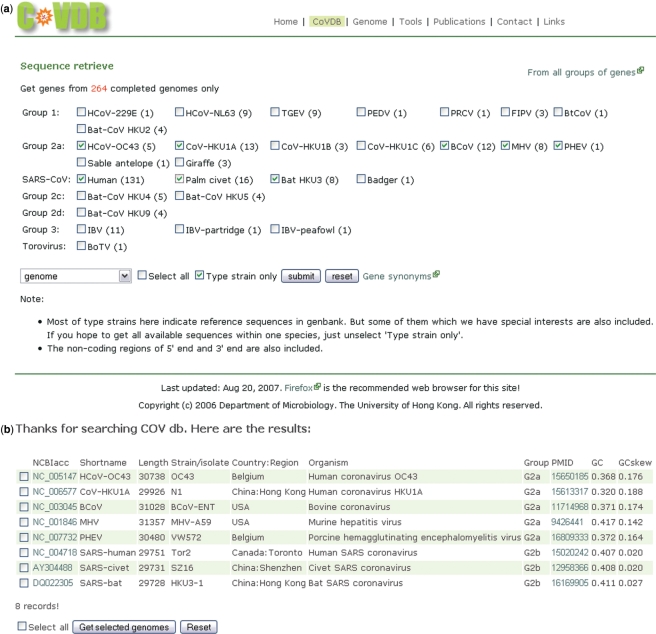
The main goal for setting up CoVDB is to provide a convenient and efficient platform for retrieving batches of coronavirus gene sequences. The interfaces of the database are simple and user friendly. All genes and genomes contain links to GenBank and/or pubmed. CoVDB contains two main pages for sequence retrieval. From the homepage, one can enter the first main page for retrieval of complete genomes and their genes by clicking ‘CoVDB’ (Figure 2a). From this page, users can obtain genes from specific coronavirus species by selecting the corresponding check boxes. We defined one representative genome from each species as the ‘Type strain’. Most of the time, this ‘Type strain’ is the one assigned as the reference sequence in GenBank. By choosing the ‘Type strain only’ option, users can obtain one gene sequence per species and construct phylogenetic tree or perform other comparisons. An example of retrieving complete genome or a specific gene of complete genome of selected species is shown in Figure 2b and c.
Figure 2.
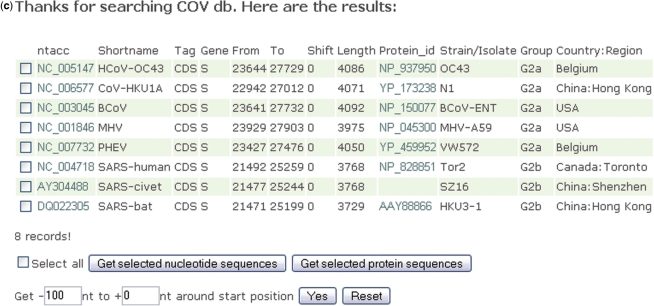
Screenshots of CoVDB complete genome retrieval pages. (a) Specific gene can be retrieved using the pull-down list at the left lower corner. The number in brackets indicates the number of complete genomes for that coronavirus. (b) Example of showing genomes of selected species (some group 2a coronaviruses and SARS-CoV-related coronaviruses). Default is to show the ‘Type strain’ for each species only. The columns NCBIacc and PMID link to GenBank and pubmed, respectively. (c) Example of showing S gene of selected species by choosing S in the pull-down list. For genes downstream to orf1ab, sequences upstream to the initiation codons can also be retrieved from this result page. This function is particularly useful for the detection of transcription regulatory sequences.
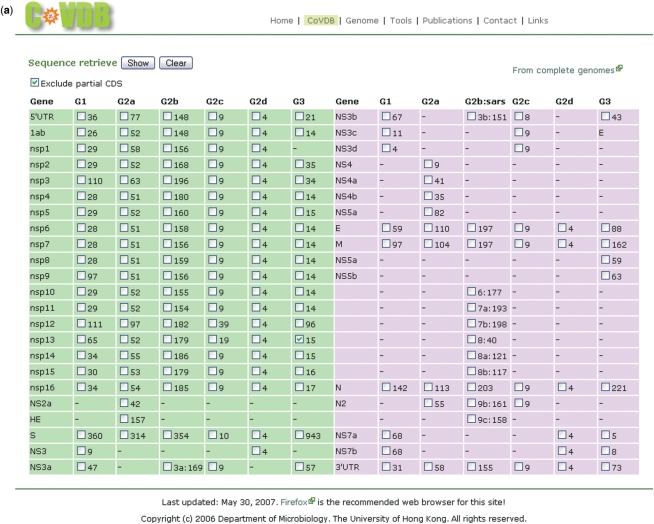
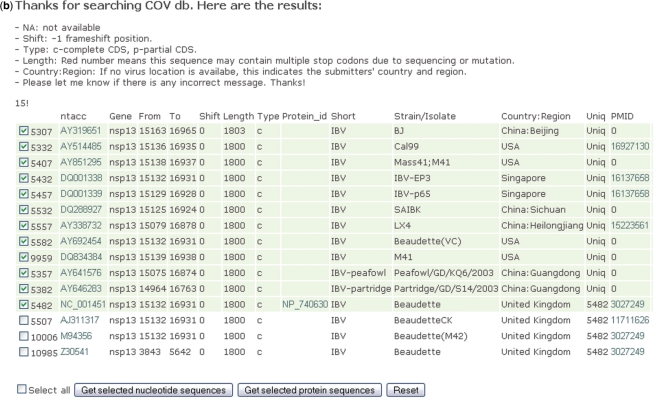
From the page for retrieval of complete genomes and their genes, one can enter the second main page for retrieval of all complete and/or incomplete genes of a coronavirus (Figure 3a) by clicking ‘From all groups of genes’. In this page, all the gene sequences are grouped vertically according to which coronavirus group and subgroup they belong to, and horizontally by the names of the genes. The option ‘Exclude partial CDS’ can be used if only complete genes are required. An example of retrieving all the sequence of a particular gene for a group of coronavirus is shown in Figure 3b. If the translated sequence of a selected gene has more than one stop codon which is probably due to sequencing error, the number in the ‘Length’ column of this gene will be marked in red.
Figure 3.
Screenshots of all gene retrieval pages. (a) Gene sequences are grouped vertically according to which coronavirus group and subgroup they belong to, and horizontally by the name of the genes. The numbers next to each checkbox indicates the number of that gene in CoVDB. The option ‘Exclude partial CDS’ can be used if only complete genes are required. (b) Example of showing the 15 sequences of nsp13 in group 3 coronaviruses. The first column is CoVDB gene id. In the Uniq column, ‘Uniq’ will be shown if there is no other identical sequence in CoVDB. Otherwise, gene id of the sequences identical to it will be shown.
Polyprotein annotation
In all coronavirus genomes, orf1ab occupies two-thirds of the genome and it is translated as a polyprotein. This polyprotein is post-translationally cleaved by 3C-like protease (3CLpro) and papain-like protease (PLpro) into 15–16 non-structural proteins. Some of the non-structural proteins, such as RNA-dependent RNA polymerase, helicase, 3CLpro and PLpro are essential for replication or virulence of the coronavirus, although the functions of others are still unclear. Due to the essentiality of the non-structural proteins, these sequences are often used for evolutionary analysis, primer design, etc. However, except for the reference sequences, detailed cleavage site information is not provided for the non-structural proteins in other sequences in GenBank. Since it has been shown that 3CLpro and PLpro of coronavirus cleave at conserved specific amino acids, the putative cleavage sites of the 15–16 non-structural proteins can be predicted by multiple sequence alignment. Using these pieces of information, we have annotated these non-structural proteins in all the coronavirus sequences for easy retrieval in CoVDB.
Protein/gene name unification
By convention, all non-structural proteins in the polyprotein encoded by orf1ab are named as ‘nsp’, with each protein numbered consecutively starting from the 5′ end (nsp1–nsp16). The structural proteins after the polyprotein are hemagglutinin esterase (HE, in group 2a coronaviruses), spike glycoprotein (S), envelope protein (E), membrane protein (M) and nucleocapsid protein (N). However, there is no unified naming system for the non-structural proteins encoded by ORFs downstream to orf1ab. This lack of a unified system greatly reduces the stability and accuracy of ortholog retrieval.
In CoVDB, with the aim of facilitating gene retrieval, we tried to unify the naming of these non-structural proteins from different groups of coronaviruses. On the other hand, we have also tried to avoid radical changes in the names that may lead to confusion. In CoVDB, these non-structural proteins are named as NS2a, NS3x, NS4x, NS5x and NS7x (x = a, b, c,…). NS2a denotes the ORF between orf1ab and HE of group 2a coronaviruses. NS3x denotes the ORFs between S and E of groups 1, 2c, 2d and 3 coronaviruses. In most of these coronaviruses, there are two NS3x, named NS3a and NS3b. However, in group 1 coronaviruses, the genomes of some members (e.g. HCoV-NL63, PEDV) contain only one ORF between S and E. When we compared their putative amino acid sequences to the corresponding ones in other group 1 coronavirus genomes using BLAST, as well as searching for conserved domains using motifscan, results showed that the putative proteins encoded by these ORFs belonged to a protein family in Pfam originally assigned as ‘Corona_NS3b’ (accession number PF03053). Therefore, we named these ORFs as NS3b. NS4x denotes the ORFs between S and E of group 2a coronaviruses. NS5x denotes the ORFs between M and N of group 3 coronaviruses. One exception is NS5a of group 2a coronaviruses. Traditionally, this name denotes an ORF upstream of E in group 2a coronaviruses. Therefore, we have kept this name for that ORF in CoVDB. NS7x denotes the ORFs downstream of N gene. It is important to note that due to variations in genome organizations among different groups of coronaviruses (Table 1), NS genes with the same name in different coronavirus groups may not be orthologs of each other. The complete genome gene search page of CoVDB contains a link to a Gene synonyms page, which includes a list of synonymous names of the various genes in the coronavirus genomes.
Table 1.
Genome organization of different groups of coronavirus
| Group | Organizations |
|---|---|
| 1 | 5′UTR-nsp1-16-S-NS3x-E-M-N-(NS7x)-3′UTR |
| 2a | 5′UTR-nsp1-16-(NS2a)-HE-S-(NS4x)-NS5a-E-M-N-3′UTR |
| 2b | 5′UTR-nsp1-16-S-sars3x-E-M-sars6-sars7x-sars8x-N-3′UTR |
| 2c | 5′UTR-nsp1-16-S-NS3x-E-M-N-3′UTR |
| 2d | 5′UTR-nsp1-16-S-NS3x-E-M-N-(NS7x)-3′UTR |
| 3 | 5′UTR-nsp1-16-S-NS3x-E-M-NS5x-N-(NS7x)-3′UTR |
Identical sequence labeling
Sequence redundancy is another problem of coronavirus sequences in public nucleotide databases. Different strains of the same species from samples collected in different locations or at different times may possess completely or partially identical sequences. These sequences, though containing important epidemiological information, increase the workload during sequence analysis. In CoVDB, we compared all nucleotide sequences and labeled the identical ones to mitigate this problem. Users can choose to show or not to show strains with identical sequences by clicking on the check boxes to the left of the page (Figure 3b).
Blast similarity search
During the process of coronavirus gene sequences analysis, we encountered a major problem when coronavirus gene sequences, especially those of orf1ab, were used for blast search against GenBank or any other coronavirus databases. When part of the orf1ab gene (e.g. nsp5) is used as the query sequence, instead of getting the gene for the specific non-structural protein that the query sequence is homologous to, the results will only show that the hits are within orf1ab, or in some cases, shown to be within the entire coronavirus genome. Much time will be needed for further analyzing the results manually in order to locate the positions of the cleavage sites of the corresponding genes for the non-structural proteins, making it very inefficient for further downstream work.
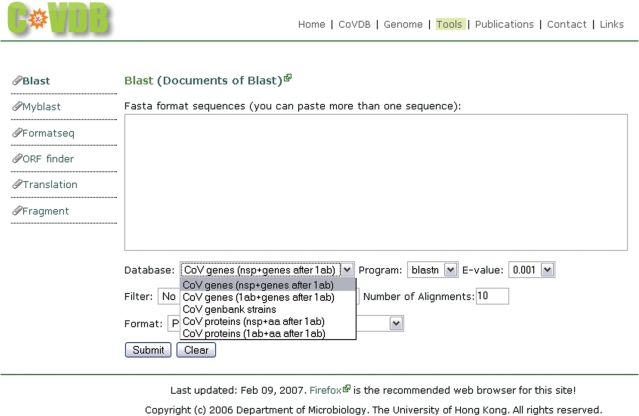
This problem has been overcome by the annotated sequences in CoVDB. The blast search page of CoVDB is an interface for facilitating coronavirus similarity search. The background support program, blastall, is from the NCBI Blast package. The blast search page can be entered by clicking ‘Tools’ in the top menu bar in any page of CoVDB. Since all sequences in CoVDB are annotated, they can be grouped into different datasets for blast search. Users can choose one of the three nucleotide and two protein sequence datasets as the database for comparison (Figure 4). The three nucleotide sequence datasets are: CoV genes (nsp + genes after 1ab), CoV genes (1ab + genes after 1ab) and CoV GenBank strains, which are the original sequences retrieved from GenBank. The two protein sequence datasets are the translated sequences of the first two nucleotide datasets: CoV proteins (nsp + aa after 1ab) and CoV proteins (1ab + aa after 1ab).
Figure 4.
Screenshot of blast similarity search page. Five datasets can be chosen as the database for comparison.
MyBlast
‘MyBlast’ employs the same blast program as the Blast page mentioned above. However, instead of selecting a predefined nucleotide or amino acid sequence database, multiple sequences can be pasted into the second sequence input box to generate a temporary sequence database. One or more query sequences can be pasted into the first sequence input box for blastn or blastp search against the temporary sequence database.
ORF finder for coronavirus
This ORF finder is specifically designed for coronavirus genome analysis. The result page shows the positions and lengths of each putative ORF and the position of the putative ribosomal frameshift site for translation of orf1ab. The nucleotide or amino acid sequences of the ORFs can be shown by selecting the corresponding check boxes. To facilitate genome comparison and annotation, the most closely related coronavirus, which had been annotated in CoVDB, can be chosen from a pull-down list for comparison using blast search. This function is particularly useful for determining the range of nsp in orf1ab.
DISCUSSION
Rapid and accurate batch sequence retrieval is both the cornerstone and bottleneck for comparative gene or genome analysis. During the process of complete genome sequencing and comparative analysis of the various novel human and animal coronavirus genomes in the past 2 years, we have developed a comprehensive database, CoVDB, of annotated coronavirus genes and genomes, which offers efficient batch sequence retrieval and analysis. As shown by our experience in using CoVDB for comparative genome analysis of novel coronaviruses we have discovered (4,13,16,18,19), we find that CoVDB is more rapid and efficient than other existing coronavirus databases for batch sequence retrieval for the following reasons. First, we have performed annotation on all non-structural proteins in the polyprotein encoded by orf1ab of every single sequence. Second, annotation was performed for the non-structural proteins encoded by ORFs downstream to orf1ab using a standardized system, with some exceptions given to some names that have been used for a long time so as to minimize confusion. Third, all sequences with identical nucleotide sequences were labeled where one can choose to show or not to show strains with identical sequences. Fourth, CoVDB contains not only complete coronavirus genome sequences, but also incomplete genomes and their genes. Some genes of coronaviruses, such as pol, spike and nucleocapsid are sequenced much more frequently than others because they are either most conserved or least conserved. These gene sequences are particularly important for evolutionary analysis, single nucleotide polymorphism studies and design of primers for RT-PCR or quantitative RT-PCR amplification.
Availability
CoVDB is constructed by the Department of Microbiology, the University of Hong Kong. It is available at no charge at http://covdb.microbiology.hku.hk.
ACKNOWLEDGEMENTS
We are grateful to the generous support of Mr Hui Hoy and Mr Hui Ming in the genomic sequencing platform. This work is partly supported by the Research Grant Council Grant; University Development Fund and Outstanding Young Researcher Award, The University of Hong Kong; The Tung Wah Group of Hospitals Fund for Research in Infectious Diseases; the HKSAR Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau; and the Providence Foundation Limited in memory of the late Dr Lui Hac Minh. Funding to pay the Open Access publication charges for this article was provided by Research Grant Council Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brian DA, Baric RS. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai MM, Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ziebuhr J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 2004;7:412–419. doi: 10.1016/j.mib.2004.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo PC, Lau SK, Yip CC, Huang Y, Tsoi HW, Chan KH, Yuen KY. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 6.Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, Khattra J, Asano JK, Barber SA, et al. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 7.Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 9.Woo PC, Lau SK, Tsoi HW, Chan KH, Wong BH, Che XY, Tam VK, Tam SC, Cheng VC, et al. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet. 2004;363:841–845. doi: 10.1016/S0140-6736(04)15729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouchier RA, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC, Simon JH, Osterhaus AD. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl Acad. Sci. USA. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, et al. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo PC, Huang Y, Lau SK, Tsoi HW, Yuen KY. In silico analysis of ORF1ab in coronavirus HKU1 genome reveals a unique putative cleavage site of coronavirus HKU1 3C-like protease. Microbiol. Immunol. 2005;49:899–908. doi: 10.1111/j.1348-0421.2005.tb03681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, Wong BH, Poon RW, Cai JJ, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo PC, Lau SK, Tsoi HW, Huang Y, Poon RW, Chu CM, Lee RA, Luk WK, Wong GK, et al. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J. Infect. Dis. 2005;192:1898–1907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo PC, Lau SK, Li KS, Poon RW, Wong BH, Tsoi HW, Yip BC, Huang Y, Chan KH, et al. Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau SK, Woo PC, Li KS, Huang Y, Wang M, Lam CS, Xu H, Guo R, Chan KH, et al. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007 doi: 10.1016/j.virol.2007.06.009. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang XC, Zhang JX, Zhang SY, Wang P, Fan XH, Li LF, Li G, Dong BQ, Liu W, et al. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo PC, Wang M, Lau SK, Xu H, Poon RW, Guo R, Wong BH, Gao K, Tsoi HW, et al. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 2007;81:1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavanagh D, Mawditt K, Welchman Dde B, Britton P, Gough RE. Coronaviruses from pheasants (Phasianus colchicus) are genetically closely related to coronaviruses of domestic fowl (infectious bronchitis virus) and turkeys. Avian Pathol. 2002;31:81–93. doi: 10.1080/03079450120106651. [DOI] [PubMed] [Google Scholar]
- 21.East ML, Moestl K, Benetka V, Pitra C, Honer OP, Wachter B, Hofer H. Coronavirus infection of spotted hyenas in the Serengeti ecosystem. Vet. Microbiol. 2004;102:1–9. doi: 10.1016/j.vetmic.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonassen CM, Kofstad T, Larsen IL, Lovland A, Handeland K, Follestad A, Lillehaug A. Molecular identification and characterization of novel coronaviruses infecting graylag geese (Anser anser), feral pigeons (Columbia livia) and mallards (Anas platyrhynchos) J. Gen. Virol. 2005;86:1597–1607. doi: 10.1099/vir.0.80927-0. [DOI] [PubMed] [Google Scholar]
- 23.Liu S, Chen J, Chen J, Kong X, Shao Y, Han Z, Feng L, Cai X, Gu S, et al. Isolation of avian infectious bronchitis coronavirus from domestic peafowl (Pavo cristatus) and teal (Anas) J. Gen. Virol. 2005;86:719–725. doi: 10.1099/vir.0.80546-0. [DOI] [PubMed] [Google Scholar]
- 24.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snyder EE, Kampanya N, Lu J, Nordberg EK, Karur HR, Shukla M, Soneja J, Tian Y, Xue T, et al. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 2007;35:D401–D406. doi: 10.1093/nar/gkl858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]