Abstract
Alternative splicing is known to be one of the major sources for functional diversity in higher eukaryotes. Several splicing isoforms have been characterized in the literature that play important roles in cellular processes like apoptosis or signal transduction pathways. Splicing events can often be detected on the mRNA level by large-scale cDNA or EST experiments and such data is collected and annotated in several databases. Nevertheless, the effects of splicing on the structure of a protein are largely unknown. The ProSAS (Protein Structure and Alternative Splicing) database fills this gap and provides a unified resource for analyzing effects of alternative splicing events in the context of protein structures. ProSAS comprehensively annotates and models protein structures for several Ensembl genomes as well as SwissProt entries harbouring splicing events. Alternative isoforms annotated in Ensembl or SwissProt can be analyzed on the protein structure and protein function level using an intuitive user interface that provides several features and tools for a structure-based analysis of alternative splicing events. The ProSAS database is freely accessible at http://www.bio.ifi.lmu.de/ProSAS.
INTRODUCTION
Alternative splicing assembles the exons of a gene in different ways during pre-mRNA splicing, such that different transcripts are produced from the same genomic locus (gene). Based on EST data, it is estimated that up to 74% of the human multi-exon genes are alternatively spliced (1). Therefore, splicing largely increases the number of possible gene products in the human proteome and in correspondence with time and tissue-specific regulation of alternative splicing increases its functional complexity.
Several databases published in recent years provide access to alternative splicing data as well as features for a functional characterization of the different isoforms. Among them are the databases ASD (2), ASAP II (3), H-DBAS (4), ECgene (5), FAST-DB (6) and ASTALAVISTA (7). Most databases (ASD, ASAP II, ECgene and H-DBAS) are mainly dedicated to the collection of alternative transcripts on the mRNA level. Splicing isoforms are annotated with InterPro (8) patterns, tissue specificity of transcripts and literature describing specific isoforms. Though such data are a very valuable source of information, one dimension of the divergence of the different isoforms, namely the protein structure, is missing.
The effects of alternative splicing onto protein structures are not very well understood also due to missing experimental data of splicing isoforms in the PDB (9). As several bioinformatics studies (10–12) have shown, the effects of alternative splicing onto protein structures are in many cases non-trivial. Therefore, a structure-based analysis of splicing isoforms is necessary in order to understand the possible functional role of an isoform in the cell. ProSAS unifies protein structure and alternative splicing data for several mammalian genomes and provides tools and data for a detailed analysis of splicing events on the protein structure level.
THE PROSAS DATABASE CONTENT AND METHODS
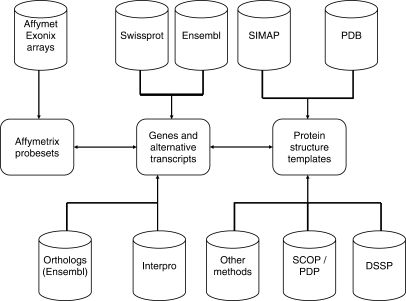
Figure 1 shows an overview on the database content and the annotation pipeline used in the ProSAS database, while Table 1 summarizes the current content of the database.
Figure 1.
Database content and annotation pipeline. Gene and alternative transcript information is obtained from the Ensembl database as well as from SwissProt. Additional information comes from InterPro patterns and orthologs are mapped by Ensembl database using BioMart. Protein structures from the PDB are annotated to transcripts using SIMAP and several structural features are computed using SCOP, PDP and DSSP annotations. Other data like the (structural) variance in the corresponding protein family will be added in the future (Other methods). Affymetrix probesets are mapped onto transcripts and genes.
Table 1.
Summary of the content of the ProSAS database.
| Genes | Transcripts | Exons | Structurally modelled transcripts (genes) | ||
|---|---|---|---|---|---|
| Seqid > 0.4, cov > 0.75 | seq_id > 0.4 | ||||
| Human | 26 228 | 50 539 | 256 257 | 7601 (4504) | 18 520 (9433) |
| Mouse | 24 423 | 32 041 | 205 865 | 4912 (4167) | 10 673 (8330) |
| Rat | 23 265 | 33 657 | 219 304 | 5348 (4123) | 11 680 (8229) |
| Total | 73 916 | 11 6237 | 681 426 | 17 861 (12794) | 40 873 (25 992) |
| Proteins | Isoforms | Modelled SwissProt proteins | |||
| SwissProt | 12 530 | 33 155 | 1949 | 5767 | |
The last two columns give information about the coverage of human, rat and mouse genes and transcripts as well as SwissProt proteins, respectively, with respect to two different criteria. The first column requires a sequence identity between target and template of at least 40% (save modelling zone) and a structural coverage of the transcript (SwissProt protein) sequence of 75%. The second column displays all transcripts and genes (SwissProt proteins) that have at least one template assigned with a sequence identity larger than 40%. In total, the database covers about 17% of the genes and 15% of the SwissProt proteins with high quality, full length models and about 35% of the genes and 46% of all SwissProt entries with high quality, partial structures.
Genome and alternative splicing data
Genomic data available in ProSAS is based on the Ensembl database (13). Currently, data for human (Homo sapiens), mouse (Mus musculus) as well as rat (Rattus norvegicus) is available. Alternative splicing events for each gene are based on the alternative transcripts annotated in Ensembl. Additionally, all SwissProt (14) entries with annotated splicing events (VARSPLIC annotation) are available in the ProSAS database. In cases where transcripts correspond to a SwissProt entry, both sources are interlinked which allows for an integrated analysis of splicing events annotated in different databases.
Protein structure data and feature assignment
Protein structure data is annotated to genes on the transcript level. Currently, we use a very strict and conservative approach to model protein structures, in order to avoid errors and misinterpretations caused by wrong structure annotations. To identify potential templates we search the SIMAP database (15) using the SIMPAT web service client for homologs of the transcript in the PDB with an e-value cut off of 10e-5. In the case that several PDB structures map to the same region of the transcript, the template with the best e-value and the largest transcript coverage is selected. For all selected templates and transcripts, we compute Smith–Waterman free-shift alignments (16) using the PAM250 matrix and gap open and extend parameters of 12 and 1, respectively. The protein structure assignments may be filtered by the user with respect to their sequence identity with the transcript and only templates with a high sequence identity of more than 40% should be used for further steps.
Structural features that help users to judge the effects of the event on the structure level are assigned to each structurally modeled gene, transcript and exon using several tools and databases. For example, secondary structure and solvent accessibility-based features are computed using DSSP (17) and structural domains are identified using the SCOP (18) database and the PDP (19) program. More features are available and discussed in the database manual.
Affymetrix data
Affymetrix provides a new type of microarray chip to measure the expression of human, mouse and rat genes on the exon level. To aid a structure-based analysis of data obtained from such expression experiments, we mapped probesets measured on the different mammalian exon array chips onto exons of human, mouse and rat, respectively. A probeset was mapped onto an Ensembl exon if the chromosomal position of the Affymetrix gene annotated in the NetAffx file overlapped at least 75% with the chromosomal position of the human gene and the position of the probeset overlapped at least 75% with the chromosomal position of the human exon.
Further data sources
Other data sources linked to genes and transcripts available in ProSAS include InterPro pattern mappings onto every exon in the database. Pattern matches of InterPro member databases were obtained from SIMAP. Orthologous genes between human, mouse and rat as well as gene descriptions are obtained from Ensembl using the BioMart (20) service.
ACCESS TO THE PROSAS DATABASE
The data stored in ProSAS are freely available for download (MySQL table dumps) or accessible through the web interface of the ProSAS database, which allows users to access and intuitively browse the data. Basically, the interface consists of three different views: a search dialog to search the database and present the results of a query as well as three types of detail views visualizing SwissProt entries or genes and transcripts, respectively.
Database searches
The ProSAS database can be searched by several identifiers: Ensembl gene and transcript ids, Ensembl gene descriptions (fulltext keyword search), genes that match certain InterPro member database patterns, SwissProt/Uniprot names and ids as well as Affymetrix probeset ids. The search may be limited to genes that are modeled by protein structure according to certain structure quality criteria. From the search results, users may proceed to the detail views.
Gene report
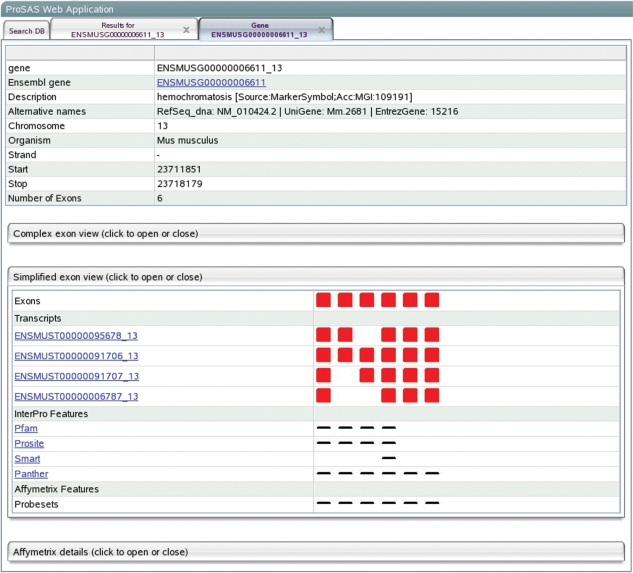
This view (Figure 2) provides access to gene-specific information. All coding exons of the gene as well as the exon composition of the corresponding transcripts of the gene are visualized as linear arrays in two subsections displaying either the exon sizes relative to their true size or in a simplified way as equally sized blocks telling if exons are present or absent in the transcript, which allows for a faster overview on annotated splicing events. Exon information (sequence, positions and phase) can be viewed by clicking on the specific exon. Links to Ensembl and SwissProt (if annotated for the gene) are provided to obtain additional information and InterPro and Affymetrix data is annotated to the exons of the gene. InterPro patterns are linked to their corresponding source databases to obtain specific information about the pattern. These patterns allow to judge splicing events in a functional context by the absence or presence of patterns in different transcripts. Following the transcript link leads to the transcript view panel.
Figure 2.
Details for gene ENSMUSG00000006611_13 from mouse showing all exons of the gene as well as different transcripts annotated for the gene in Ensembl. Matches of InterPro patterns and Affymetrix probesets onto exons are also shown.
Transcript report and structure analysis
The transcript view (Figure 3), as well as the SwissProt entry report that is designed in a very similar way, allows access to transcript specific data like the protein and DNA sequence and colors the exons of the transcript on the sequences. Other transcripts of the gene can be visualized with respect to the current transcript and deletions, replacements and insertions that occur due to the splicing event are color-coded on the transcript sequence.
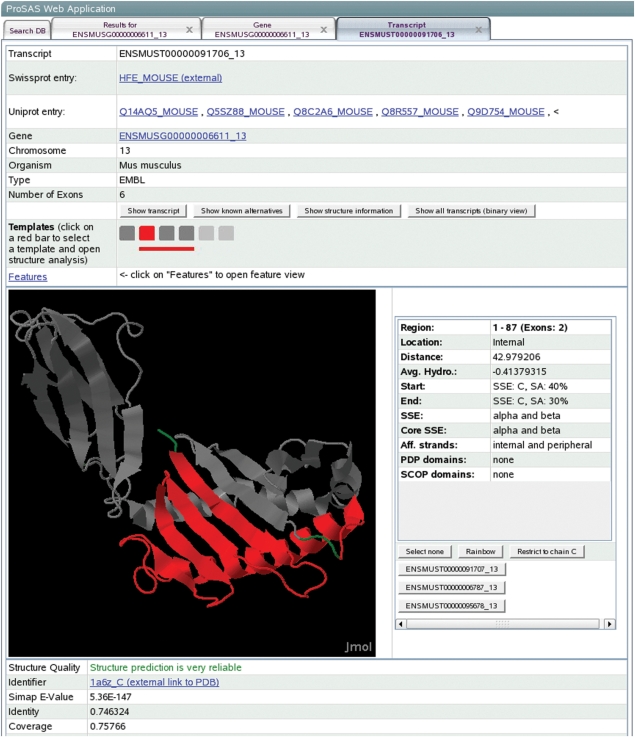
Figure 3.
Transcript details view for transcript ENSMUST00000091706_13 from gene ENSMUSG00000006611_13. The structure of the transcript is visualized with Jmol (Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/) and the difference with respect to transcript ENSMUST00000091707_13, namely the deletion of a larger N-terminal part is visualized on the structure. The alternatively spliced region is characterized with respect to different features such as solvent accessibility or secondary structure content.
The analysis of alternative splicing events in the context of protein structures, the unique feature of the ProSAS database, is also available in this view. Users can choose a protein structure that matches the transcript. This structure is then visualized using Jmol (Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org) and the correspondence of the gene structure (i.e. its exons) and the protein structure can be explored interactively. All exons of the transcript can be colored individually in the structure. That way, exon positions and substructures that belong to one or more exons on the structure level can easily be identified. Such an analysis provides useful insights into the correspondence of exons to small structural motifs or the location of exon boundaries on the structure level. An exon or a group of exons can additionally be structurally classified, in terms of many features like secondary structure content, solvent accessibility, structural contacts as well as domain classifications as defined by SCOP or PDP.
Despite analyzing each exon individually, known splicing events, i.e. other known transcripts of the gene can also be coloured on the structure level relative to the current transcript. This conveniently visualizes deletions, insertions and replacements on the structure level observed in different transcripts.
Those tools and features allow users to judge the importance of a spliced exon for the stability of a protein structure and will therefore provide an interesting additional dimension in the analysis of splicing events and isoforms.
While some events appear to be non-critical for the structure since complete domains, globular parts or unstructured regions are removed or replaced, others that remove large or well-structured parts of the protein are much harder to explain and might point to non-sense mediated mRNA decay or non-trivial effects in the isoform structure. The web interface allows identifying such cases, where additional experimental validation of the protein product might be necessary.
CONCLUSION AND FUTURE DIRECTION
With the ProSAS database, we provide a unified framework to analyze possible effects of alternative splicing in the context of the resulting protein structure. The provided data and tools allow bridging the gap between the mRNA and protein structure level, which is a critical step when trying to understand how functional diversity may arise from alternative splicing.
In the future, we plan to incorporate knowledge about protein structure evolution into the analysis of alternative splicing events. Protein structures grouped into the same family, superfamily or fold (e.g. as defined by SCOP) display insertions, replacements and deletions which were tolerated by a protein family in the course of evolution. Such ‘evolutionary isoforms’ will provide useful knowledge to judge the effects of alternative splicing events. A second direction will be to enable experimentalists using the Affymetrix exon array technology to analyze their results in the context of protein structure in an automated fashion as well as the integration of other sources for alternative splicing events into the database.
With ProSAS, we provide a useful tool for researchers trying to understand functional and structural effects of alternative splicing and we encourage users to guide and support the future development of the database with their feedback.
ACKNOWLEDGEMENTS
Funding to pay the Open Access publication charges for the article was provided by Ludwig-Maximilians-Universität (LMU) München.
Conflict of interest statement. None declared.
REFERENCES
- 1.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 2.Stamm S, Riethoven JJ, Le Texier V, Gopalakrishnan C, Kumanduri V, Tang Y, Barbosa-Morais NL, Thanaraj TA. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim N, Alekseyenko AV, Roy M, Lee C. The ASAP II database: analysis and comparative genomics of alternative splicing in 15 animal species. Nucleic Acids Res. 2007;35:D93–98. doi: 10.1093/nar/gkl884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeda J, Suzuki Y, Nakao M, Kuroda T, Sugano S, Gojobori T, Imanishi T. H-DBAS: alternative splicing database of completely sequenced and manually annotated full-length cDNAs based on H-Invitational. Nucleic Acids Res. 2007;35:D104–109. doi: 10.1093/nar/gkl854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y, Lee Y, Kim B, Shin Y, Nam S, Kim P, Kim N, Chung WH, Kim J, et al. ECgene: an alternative splicing database update. Nucleic Acids Res. 2007;35:D99–103. doi: 10.1093/nar/gkl992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de la Grange P, Dutertre M, Correa M, Auboeuf D. A new advance in alternative splicing databases: from catalogue to detailed analysis of regulation of expression and function of human alternative splicing variants. BMC Bioinformatics. 2007;8:180. doi: 10.1186/1471-2105-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foissac S, Sammeth M. ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res., 2007 July; 2007;35(Web Server issue):W297–W299. doi: 10.1093/nar/gkm311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide protein data bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl Acad. Sci. USA. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang P, Yan B, Guo JT, Hicks C, Xu Y. Structural genomics analysis of alternative splicing and application to isoform structure modeling. Proc. Natl Acad. Sci. USA. 2005;102:18920–18925. doi: 10.1073/pnas.0506770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tress ML, Martelli PL, Frankish A, Reeves GA, Wesselink JJ, Yeats C, Olason PL, Albrecht M, Hegyi H, et al. The implications of alternative splicing in the ENCODE protein complement. Proc. Natl Acad. Sci. USA. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rattei T, Arnold R, Tischler P, Lindner D, Stumpflen V, Mewes HW. SIMAP: the similarity matrix of proteins. Nucleic Acids Res. 2007;34:D252–256. doi: 10.1093/nar/gkj106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 17.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 18.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexandrov N, Shindyalov I. PDP: protein domain parser. Bioinformatics. 2003;19:429–430. doi: 10.1093/bioinformatics/btg006. [DOI] [PubMed] [Google Scholar]
- 20.Kasprzyk A, Keefe D, Smedley D, London D, Spooner W, Melsopp C, Hammond M, Rocca-Serra P, Cox T, et al. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]