Abstract
MetaCyc (MetaCyc.org) is a universal database of metabolic pathways and enzymes from all domains of life. The pathways in MetaCyc are curated from the primary scientific literature, and are experimentally determined small-molecule metabolic pathways. Each reaction in a MetaCyc pathway is annotated with one or more well-characterized enzymes. Because MetaCyc contains only experimentally elucidated knowledge, it provides a uniquely high-quality resource for metabolic pathways and enzymes. BioCyc (BioCyc.org) is a collection of more than 350 organism-specific Pathway/Genome Databases (PGDBs). Each BioCyc PGDB contains the predicted metabolic network of one organism, including metabolic pathways, enzymes, metabolites and reactions predicted by the Pathway Tools software using MetaCyc as a reference database. BioCyc PGDBs also contain predicted operons and predicted pathway hole fillers—predictions of which enzymes may catalyze pathway reactions that have not been assigned to an enzyme. The BioCyc website offers many tools for computational analysis of PGDBs, including comparative analysis and analysis of omics data in a pathway context. The BioCyc PGDBs generated by SRI are offered for adoption by any interested party for the ongoing integration of metabolic and genome-related information about an organism.
INTRODUCTION
MetaCyc (MetaCyc.org) is a non-redundant reference database of small-molecule metabolism that contains experimentally verified metabolic pathway and enzyme information curated from the scientific literature (1). Because MetaCyc contains only experimentally elucidated data, it is a unique and valuable resource. The metabolic pathways and enzymes in MetaCyc are from a wide variety of organisms with an emphasis on microbial and plant metabolism, although a significant number of animal pathways are also included.
In addition to serving as a general reference source on metabolism, MetaCyc is used in conjunction with the PathoLogic component of the Pathway Tools software (2) to predict computationally the metabolic network of any organism whose genome has been sequenced and annotated (3–5). This automated process creates the predicted network in the form of a Pathway/Genome Database (PGDB). BioCyc (BioCyc.org) is a collection of more than 350 organism-specific PGDBs resulting from the prediction of such metabolic networks using MetaCyc and PathoLogic. Computationally predicted PGDBs can subsequently be improved and updated by manual curation using the Editors component of the Pathway Tools software. Scientists can either adopt and curate existing PGDBs through the BioCyc website (biocyc.org/intro.shtml#adoption), or create their own PGDBs using MetaCyc and Pathway Tools (biocyc.org/download.shtml), which has now been done by more than 80 groups for important model organisms including Mus musculus (Mouse), Arabidopsis thaliana, Dictyostelium discoideum and Saccharomyces cerevisiae (see BioCyc.org for a more complete listing).
Pathway Tools includes a web server that enables the publishing of PGDBs either through the Internet or an internal network, and the Navigator component allows browsing and analysis of PGDBs, either locally or over the Internet. A detailed description of Pathway Tools is available at http://bioinformatics.ai.sri.com/ptools/ and (2).
PGDBs are useful in many areas of research including biochemistry, molecular biology, biotechnology, bioinformatics, metabolic engineering and systems biology. They are also useful as an educational tool.
In the past 2 years we have significantly expanded the data content of MetaCyc and BioCyc, and added major enhancements to the Pathway Tools software that supports them. These improvements are described in the following sections.
CHANGES AND ADDITIONS TO METACYC
Overview of the MetaCyc data
Data in MetaCyc are curated from the experimental literature by PhD-level curators. The pathways are curated from all kingdoms of life, with an emphasis on microbial (bacteria, archaea and fungi), higher plant (viridiplantae) and, to a lesser extent, metazoan (animal) pathways (see Figure 1 of an example for a pathway in MetaCyc). Curators at SRI cover microbial and metazoan pathways, whereas curators at the Carnegie Institution and (starting in late 2007) Dr Lukas Mueller and colleagues at Cornell University cover pathways from higher plants. The microbial portion of MetaCyc was initialized with the full complement of EcoCyc metabolic pathways (6) ensuring a comprehensive coverage of the basic pathways of central and intermediary metabolism typical of enteric bacteria. However, curation over the past few years has expanded the database significantly with pathways from a wide range of nonpathogenic bacteria, such as those responsible for nutrient cycling in the environment, detoxification of heavy metals and other environmental pollutants, and degradation of recalcitrant compounds. For example, MetaCyc covers important microbially catalyzed environmental processes such as the recycling of nitrogen (10 pathways) and sulfur compounds (37 pathways), the metabolism of single-carbon (C1) compounds (20 pathways), methanogenesis (13 pathways) and aromatic compound degradation (94 pathways). Similarly, although MetaCyc has contained most of the pathways of central metabolism in higher plants for some time, recently curated material provides an extensive coverage of plant secondary metabolism. For a comparison of MetaCyc to other metabolic pathway databases, see http://metacyc.org/MetaCycUserGuide.shtml#otherpathwaydbs.
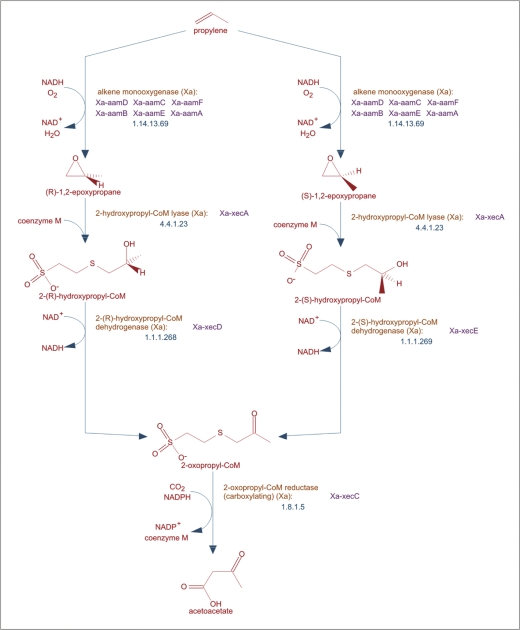
Figure 1.
An example of a pathway in MetaCyc. Pathways can be displayed at varying levels of detail. This pathway display depicts an intermediate level of detail including enzymes, EC numbers, genes and chemical structures of the main compounds. Notice the green arrow at the bottom of the pathway, which provides a hyperlink to a related downstream pathway.
Since the last Nucleic Acids Research publication 2 years ago (7), there has been a significant increase in the content of the MetaCyc database. We have added more than 400 new pathways, an increase of 57%, bringing the total number of base metabolic pathways to 977 (current statistics are taken from version 11.5 of MetaCyc, which was released on 15 August 2007). In addition, the database includes 129 superpathways (discussed later), bringing the total number of pathways to more than 1100. The number of enzymes, genes, chemical compounds and citations in the database has grown accordingly by 60, 47, 38 and 106%, respectively, and the number of organisms referenced in the database has doubled (currently at 1029), reflecting the ever-growing breadth of MetaCyc. Some database statistics are provided in Table 1.
Table 1.
The size of MetaCyc as a function of time from its first release in 1999 to the latest release in 2007 (version 11.5)
| Database objects | 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 |
|---|---|---|---|---|---|---|---|---|---|
| Metabolic pathways | 296 | 366 | 445 | 460 | 491 | 528 | 692 | 879 | 977 |
| Reactions | 3779 | 4002 | 4218 | 4294 | 4817 | 4955 | 5520 | 6113 | 6483 |
| Enzymes | 82 | 344 | 1115 | 1267 | 1543 | 1940 | 3029 | 3841 | 4332 |
| Genes | 0 | 0 | 0 | 600 | 1554 | 1821 | 2931 | 3630 | 3913 |
| Compounds | 1949 | 2180 | 2335 | 2404 | 2951 | 3551 | 4817 | 5978 | 6375 |
| Literature citations | 184 | 604 | 2381 | 2718 | 3070 | 5050 | 8599 | 11 934 | 15 199 |
Each row depicts the number of different database objects in MetaCyc during the final release for that year.
As shown in Tables 2 and 3, most of the pathways in MetaCyc occur in the bacteria and plant kingdoms, although the representation of animal pathways is increasing steadily. Because the size limit of this publication does not permit an exhaustive listing of content addition, we will provide a brief general discussion of material curated during the past 2 years.
Table 2.
List of species that have 10 or more experimentally elucidated pathways represented in MetaCyc
| Bacteria | Eukarya | Archaea | |||
|---|---|---|---|---|---|
| Escherichia coli | 225 | Arabidopsis thaliana | 189 | Methanosarcina barkeri | 18 |
| Pseudomonas putida | 40 | Homo sapiens | 79 | Sulfolobus solfataricus | 16 |
| Pseudomonas aeruginosa | 37 | Saccharomyces cerevisiae | 67 | Methanosarcina thermophila | 15 |
| Bacillus subtilis | 36 | Glycine max | 50 | Methanothermobacter thermautotrophicus | 13 |
| Salmonella typhimurium | 23 | Rattus norvegicus | 43 | Methanosarcina TM-1 | 10 |
| Pseudomonas fluorescens | 19 | Pisum sativum | 41 | ||
| Mycobacterium tuberculosis | 14 | Zea mays | 27 | ||
| Haemophilus influenzae | 13 | Solanum tuberosum | 22 | ||
| Klebsiella pneumoniae | 13 | Nicotiana tabacum | 22 | ||
| Agrobacterium tumefaciens | 13 | Oryza sativa | 20 | ||
| Deinococcus radiodurans | 12 | Spinacia oleraca | 19 | ||
| Mycobacterium smegmatis | 12 | Triticum aestivum | 16 | ||
| Sinorhizobium meliloti | 12 | Bos taurus | 16 | ||
| Paracoccus denitrificans | 11 | Hordeum vulgare | 15 | ||
| Mycoplasma pneumoniae | 11 | Lycopersicon esculentum | 15 | ||
| Acidithiobacillus ferrooxidans | 10 | Mus musculus | 15 | ||
| Cicer arietinum | 13 | ||||
| Catharanthus roseus | 12 | ||||
| Cucumis sativus | 11 | ||||
| Abies grandis | 10 | ||||
| Picea abies | 10 |
The species are grouped by taxonomic domain and are ordered within each domain based on the number of pathways (number following species name) to which the given species was assigned. Some pathways may be labeled with a higher-level taxon, such as genus, if all the species within that genus are thought to have the given pathway. However, such higher-level taxa are not included in this table.
Table 3.
Distribution of pathways in MetaCyc based on the taxonomic classification of associated species
| Bacteria | Eukarya | Archaea | |||
|---|---|---|---|---|---|
| Proteobacteria | 534 | Viridiplantae | 401 | Euryarchaeota | 60 |
| Firmicutes | 140 | Metazoa | 104 | Crenarchaeota | 26 |
| Actinobacteria | 94 | Fungi | 115 | ||
| Cyanobacteria | 22 | Euglenozoa | 11 | ||
| Bacteroidetes/Chlorobi | 21 | ||||
| Deinococcus–Thermus | 11 | ||||
| Thermotogae | 10 | ||||
| Spirochaetes | 8 | ||||
| Aquificae | 6 | ||||
| Chlamydiae–Verrucomicrobia | 5 | ||||
| Fusobacteria | 4 | ||||
| Nitrospirae | 2 | ||||
| Planctomycetes | 2 | ||||
| Thermodesulfobacteria | 2 | ||||
| Chloroflexi | 1 | ||||
| Chrysiogenetes | 1 |
Taxonomic groups (phyla for Bacteria and Archaea, kingdoms for Eukarya) are grouped by domain and are ordered within each domain based on the number of pathways (number following taxon name) associated with the taxon. Euglenozoa are listed separately as this group does not belong to any of the other eukaryotic kingdoms. A pathway may be associated with multiple organisms.
MetaCyc pathway distribution
The pathways in MetaCyc fall into four broad categories (or classes), namely, biosynthesis, degradation/utilization/assimilation, generation of precursor metabolites and energy and detoxification. During the past 2 years, we have added 192, 132, 22 and 9 pathways into these categories, respectively. The largest category is currently biosynthesis, with 530 base pathways (a base pathway is considered a lowest-level pathway in the sense that it is not subdivided into smaller component pathways). Within this category, the largest classes are secondary metabolites biosynthesis (198 pathways), amino acids biosynthesis (91), cofactors, prosthetic groups and electron carriers biosynthesis (83) and fatty acids and lipids biosynthesis (51). The second-largest category is degradation/utilization/assimilation, with 485 base pathways. Within this group, the largest classes are amino acids degradation (107), aromatic compounds degradation (94), inorganic nutrients metabolism (50), secondary metabolites degradation (48) and carbohydrates degradation (47). The third category, generation of precursor metabolites and energy, contains 100 pathways, with the largest classes being fermentation (25), respiration (17), chemoautotrophic energy metabolism (14) and methanogenesis (13). The detoxification category is substantially smaller, with only 13 pathways.
Revision and reorganization of existing MetaCyc pathways
A major emphasis over this period was placed on updating older pathways that were incompletely curated, bringing them to the current curation standards. One hundred and ninety such pathways have been extensively researched, validated and updated with current commentary, species distribution, EC reaction numbers, representative enzymes and genes and citations, reflecting the current knowledge of those pathways. Pathways that failed validation, or were deemed redundant, were deleted from the database.
Over the past 2 years we refined our criteria for defining base-pathway boundaries to minimize redundancy in the database (8). We frequently observe cases where multiple metabolic pathways share a common subsequence of reaction steps. For example, dozens of different aromatic compounds are processed by different enzymes to the common intermediate 2-oxopentenoate, which is then processed in three enzymatic steps to acetyl-CoA, an intermediate of central metabolism.
Rather than defining long pathway database objects, whose final steps after the common intermediate duplicate a shared set of reactions, we now define one pathway containing the shared set of reactions, beginning at the common intermediate, and we define separate pathways for each set of reactions that lead to the common intermediate. We use our previously introduced ‘pathway links’ to link each pathway that leads to the shared intermediate to the single pathway that consumes that intermediate. Using this methodology, we avoid repeating common steps in hundreds of pathways, and decrease substantially the redundancy in MetaCyc. To enable a larger view of the metabolic network, we use superpathways, which are larger pathways created by connecting two or more base pathways. The superpathways are treated differently than base pathways by our software, and do not contribute to redundancy. During the past 2 years we increased the number of superpathways by 100%, from 53 to 106.
Curation of taxonomic domains for MetaCyc pathways
MetaCyc is used as a reference database for pathway predictions in new organisms, as part of the creation of a new PGDB by the PathoLogic software. An undesired phenomenon we observed is the incorrect prediction of certain pathways that occurs because certain enzymes can participate in several pathways. The fact that those enzymes are present in an organism can mislead the software into predicting that all pathways associated with these enzymes are valid, when, in fact, only a portion of them really exist. To help alleviate this problem, we added taxonomic range data to every pathway in MetaCyc. The software uses this information to decrease the occurrences of such false-positive predictions by requiring a higher level of evidence to predict the presence of a pathway outside of its taxonomic domain. For example, a higher degree of evidence is required to predict, in a bacterium, the presence of a pathway whose taxonomic domain is specified as ‘plants’.
Addition of electron transport reactions
We recently enhanced the schema and user interface of MetaCyc and other PGDBs by adding the ability to represent electron transport reactions (ETRs). These reactions are used to represent the electron transport processes that occur in membrane-associated enzyme complexes, involving membrane-bound electron carriers.
ETRs are defined as a combination of two or more redox half-reactions that include such information as redox potential and the compartmental location of electrons. A new graphical diagram for an ETR visually conveys the key features of such processes, such as the direction of the electron flow and the cell-compartment locations where the substrates are transformed. The system supports both scalar and vectoral ETRs.
Author credit system
To ensure that authors of certain MetaCyc objects (pathways and enzymes) receive full credit for their work, and to enable tracking of the history of such objects, we introduced a new author crediting system. The new system, which credits both individuals and organizations, can track different types of contributions, including creation, review and revision, along with time stamps. When a pathway is transferred from one PGDB to another by the pathway import/export facility, the author credits are transferred along. For example, when one of our users submits a pathway to MetaCyc, the author credits will be transferred along and show up on the MetaCyc web server pages.
Enzyme commission updates added to MetaCyc
MetaCyc is routinely updated with data from the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB), which includes new and modified EC numbers. The last supplement that has been incorporated is supplement 11 (http://www.chem.qmul.ac.uk/iubmb/enzyme/supplements/sup2005/); supplements 12 and 13 should be incorporated by the time this article is published.
Software improvement
During the past 2 years we introduced many non-data-related improvements and additions to MetaCyc. Although these additions are far too numerous to recount here, the following paragraph lists a number of additions that directly affect the usability of the software and are visible to our users.
Compound display pages
Compound display windows now list those enzymes that are activated or inhibited by the compound, and those enzymes that require the compound as a cofactor.
Gene-reaction schematics
Gene-reaction schematics connect genes, enzymes and the reactions they catalyze. As more and more enzymes are curated into MetaCyc, the diagrams can become complex, especially when enzymes catalyze multiple reactions. In MetaCyc, these diagrams are now broken into different sections for each organism, greatly simplifying the displays.
Google searching
The MetaCyc query page now contains a section for performing a text-based search of the PGDB. This allows more flexible searching, such as phrases in pathway summaries or author names. The search uses Google's index of the PGDB.
New Search-All box
A ‘Search-All’ box is now present at the bottom of every MetaCyc web page to allow users to perform a new search without having to go back to the query page.
Mouse-over popup windows
Hovering the mouse over compounds, genes and proteins activates popup windows with additional information about the objects. The feature is now active in both the web server and the desktop versions.
THE BIOCYC DATABASE COLLECTION
A major application of MetaCyc is its use in the prediction of the metabolic pathways of an organism from its sequenced genome (9–11) using the PathoLogic program (12). BioCyc is a collection of organism-specific PGDBs (13) created by PathoLogic. Most BioCyc PGDBs were created by SRI, but some were contributed to BioCyc from Pathway Tools users outside of SRI.
The BioCyc PGDBs were created through an automated computational pipeline described below. Based on the amount of subsequent manual review and updating they received, the BioCyc databases are organized into three tiers.
Tier 1 PGDBs were created through intensive manual efforts, and receive continuous updating.
Tier 2 PGDBs have undergone moderate amounts of review and updating.
Tier 3 PGDBs received no subsequent manual review or updating.
In the past 2 years, the number of BioCyc PGDBs has substantially increased from 204 to 371 (version 11.5), out of which two are in Tier 1 (EcoCyc and MetaCyc), 20 are in Tier 2 and the rest belong to Tier 3. All the Tier 3 PGDBs and some of the Tier 2 PGDBs are available for adoption by biologists under an open-license agreement. We believe that the adoption of PGDBs for ongoing curation and updating by experts in the field will greatly facilitate the enormous task of creating up-to-date knowledge resources for each and every organism with a sequenced genome.
Methodology for creation of BioCyc databases
Here, we summarize the methodology used by SRI to create the BioCyc databases. This methodology does not apply to those BioCyc PGDBs that were contributed to BioCyc by outside groups. To obtain information about the mechanism by which a particular BioCyc PGDB was created, go to the BioCyc query page (http://biocyc.org/server.html), select the PGDB of interest and click on Organism Summary.
The input to the SRI computational pipeline used to create BioCyc PGDBs is the annotated genome for an organism M, as obtained from the CMR database (14). In the vast majority of cases, we use the annotation performed by the original sequencing center that sequenced the genome, under the rationale that this annotation is likely to involve significant manual oversight of a computational genome annotation pipeline by annotation experts, and is thus likely to be more accurate than a purely automated genome annotation. However, in a minority of cases we have observed genome annotations in which very few genes were assigned to specific functions. In such cases, we select the automatic genome annotation performed by the CMR group. The annotation used is indicated in the BioCyc organism summary page, as described in the previous paragraph.
The BioCyc computational pipeline performs the following operations:
Every gene, protein and RNA gene product described in the input genome annotation is instantiated as a database object in the PGDB.
Metabolic pathways are predicted in the PGDB by PathoLogic (12). Each predicted metabolic pathway is annotated with a computational evidence code.
Operons are predicted in the PGDB by PathoLogic (13). Each predicted operon is annotated with a computational evidence code.
Pathway hole fillers are predicted in the PGDB by PathoLogic (15). A pathway hole is a reaction in a metabolic pathway to which no enzyme has been assigned. The pathway hole filler program identifies in the genome candidate genes (‘hole fillers’) whose function is to catalyze such reactions. Each predicted pathway hole filler contains a descriptive history note on the BioCyc protein page that indicates the putative enzymatic function assigned to this protein. This is the only case in which new gene functions are inferred by SRI's computational pipeline.
Transport reactions are inferred for transporters by a PathoLogic module that analyzes the textual descriptions of transporter functions in the genome annotation. For example, a protein whose function is annotated as ‘ABC transporter for l-lysine’ in a Gram-negative bacterium is interpreted as the reaction: l-lysine [periplasm] + ATP = l-lysine [cytoplasm] + ADP + Pi. That is, the transporter moves l-lysine from the periplasm to the cytoplasm in conjunction with hydrolysis of ATP. The reaction is created in the PGDB, and is associated with the transporter.
A metabolic map diagram suitable for both on-screen display and printing as a high-resolution poster is computed for the PGDB.
The Tier 3 BioCyc PGDBs will be regenerated on a regular basis to take advantage of improvements to MetaCyc and PathoLogic. In the past, the entire collection was regenerated at once, every 6–9 months. In the future, we hope to perform more frequent regenerations. Each organism summary page identifies when a given PGDB was generated.
ENHANCEMENTS TO THE PATHWAY TOOLS SOFTWARE AND BIOCYC WEBSITE
The Pathway Tools software provides query and visualization services for MetaCyc and BioCyc. The software can run as a web server-in this mode it powers the BioCyc website. It can also run as a desktop application. Platforms supported for both modes are PC/Windows, PC/Linux (32-bit and 64-bit) and Sun/Solaris. We expect to support the Macintosh sometime in 2008. Although most capabilities of the software are present in both Web and desktop modes, some features are present in one mode only. For example, most comparative analysis operations are provided in Web mode only, but visual comparison of metabolic map diagrams is provided in desktop mode only.
SRI has developed several analysis tools that can be used with the PGDBs in the BioCyc collection. These tools are available on the BioCyc website and/or through a desktop installation of Pathway Tools, and include different viewers, comparative analysis, metabolite tracing and a set of Omics Viewers-tools for painting omics datasets (e.g. gene and protein expression and metabolomics data) onto different diagrams that can show the full genome, metabolic network or regulatory network of an organism.
During the past 2 years we have expanded the functionality of Pathway Tools in the following respects.
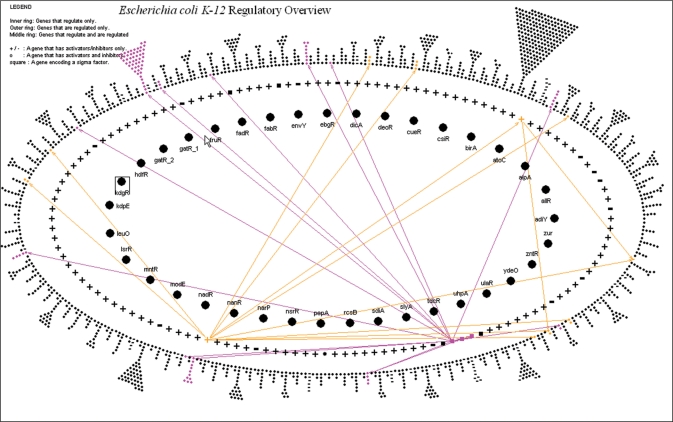
Regulatory overview
This tool displays the transcriptional regulatory network of an organism that is defined in a PGDB (Figure 2). The network can be queried in several ways, for instance by highlighting all genes under a specified Gene Ontology class or all genes regulated by a specified transcription factor. The Regulatory Overview is currently available in desktop mode only and will be available through the Web in a later release of Pathway Tools.
Figure 2.
The regulatory overview. The diagram is composed of three nested ellipses. The innermost ellipse comprises genes that are regulating other genes, but that are not regulated by any genes. The middle ellipse comprises genes that are both regulators and regulatees (= regulated by some entity), and the outermost ellipse comprises genes that are regulated, but do not regulate. The triangles that extend outward from the outer ellipse are collections of many genes that share the same set of regulator genes-although genes within a triangle may respond to those regulators in different ways. They are drawn within triangles simply to keep the size of the outer ellipse manageable.
The operation of the Regulatory Overview depends on the presence of the organism's transcriptional regulatory information within the PGDB. Currently, EcoCyc is the only BioCyc PGDB that contains this information. PGDB authors can define such a network manually using the interactive editors within Pathway Tools.
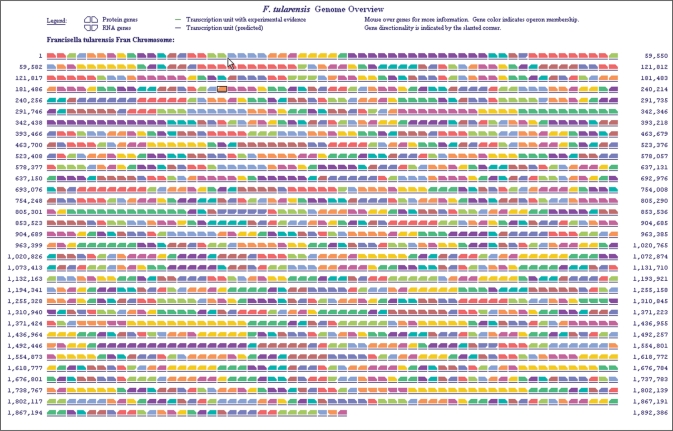
Genome overview
This tool (Figure 3) provides a one-screen view of every gene on one or more replicons (chromosomes and plasmids), and can display omics data across those entire replicons.
Figure 3.
The genome overview. This tool allows the user to view the full chromosome of an organism on a single-page diagram, and to paint expression or other omics data onto it.
Tracks in genome browser
We enhanced the Pathway Tools genome browser by adding the ability to show tracks, in a manner similar to other genome browsers. Tracks allow genome regions defined in a GFF input file to be graphically highlighted in the genome browser.
Metabolite tracing
A new metabolite tracing tool allows the user to visually trace the path of substrates through the metabolic network within a PGDB, using the Cellular Overview diagram. A sample image is available at http://biocyc.org/metab-trace.gif.
BioVelo query language
We recently introduced a new advanced query language for constructing complex SQL-like queries to PGDBs in a graphically intuitive fashion, called BioVelo. BioVelo replaces the old Advanced Query Page. Users can construct BioVelo queries interactively through the BioCyc Advanced Query Page available at http://biocyc.org/query.html.
Gene ontology support
Pathway Tools now provides the ability to display and edit assignment of Gene Ontology terms to genes within a PGDB.
Generation of metabolic map posters and genome posters
Pathway Tools can now generate two types of diagrams that are suitable for printing in a large poster format. The diagrams are a high-detail version of the cellular overview diagram (metabolic map) (http://nar.oxfordjournals.org/cgi/data/34/13/3771/DC1/2), and a genome map diagram (an online example of the diagram is provided as supplementary data for this article). These posters can be generated from any PGDB. This enhancement is available in the desktop mode only.
Sequence retrieval tool
This new tool allows the user to retrieve regions of nucleic acid sequence for a given replicon in any PGDB.
Schema updates
We have recently upgraded the Pathway Tools schema to allow representation of signaling interactions and of many types of cellular regulatory information.
Reaction editor
A complete reimplementation of the reaction editor allows the user to enter not only metabolic and transport reactions, but also Redox half reactions, electron transfer reactions and reactions involved in signal transduction.
Consistency checker
This new set of tools performs consistency checking and computation of cached data for a PGDB. The consistency checking tools search for many common types of malformed data within a PGDB, and report their findings through a graphical interface. In many cases the tools repair these malformed data automatically.
Automatic patch loading
Upon starting up, Pathway Tools now checks for available software patches, and if any are found, automatically downloads them through the Internet and installs them.
Many other enhancements to Pathway Tools are described at http://bioinformatics.ai.sri.com/ptools/release-notes.html.
HOW TO LEARN MORE ABOUT METACYC AND BIOCYC
Additional information about MetaCyc and BioCyc is available in several formats. Our websites include many informational pages, including an online guided tour of BioCyc (http://biocyc.org/samples.shtml) and a user guide for MetaCyc (http://www.metacyc.org/MetaCycUserGuide.shtml). Webinar videos that combine narration and practical demonstration of different topics can be downloaded from http://biocyc.org/webinar.shtml or from the iTunes store. We routinely publish our work in peer-reviewed journals, and a list of publications is available at http://biocyc.org/publications.shtml. In addition, we routinely host workshops that provide in-depth knowledge of our software for advanced users. To stay informed about enhancements and changes to our software, join the BioCyc mailing list available at http://biocyc.org/subscribe.shtml.
DATABASE AVAILABILITY
The MetaCyc Database, which is currently available freely for academic and nonprofit users, will become free to all users as of February 2008. The other BioCyc databases are already freely and openly available to all. See http://biocyc.org/download.shtml for download information. New versions of the downloadable data files and of the BioCyc and MetaCyc websites are released four times per year.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Veronica Armendarez for submitting data from the Streptomyces coelicolor PGDB. We thank Adam Qwan for contributions to the chemical compound data and help with quality assurance of Pathway Tools. This work was supported by grants GM70065, GM080746 and GM75742 from the NIH National Institute of General Medical Sciences. P.Z. and S.Y.R. were supported in part by NSF grant DBI-0417062, and C.T. was supported in part by a gift from Pioneer Hi-Bred. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the preceding agencies. Funding to pay the Open Access publication charges for this article was provided by the NIH National Institute of General Medical Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Krieger CJ, Zhang P, Mueller LA, Wang A, Paley S, Arnaud M, Pick J, Rhee SY, Karp PD. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2004;32:D438–D442. doi: 10.1093/nar/gkh100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karp PD, Paley S, Romero P. The Pathway Tools software. Bioinformatics. 2002;18(Suppl. 1):S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 3.Romero P, Wagg J, Green ML, Kaiser D, Krummenacker M, Karp PD. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2004;6:R2.1–R2.17. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weng S, Dong Q, Balakrishnan R, Christie K, Costanzo M, Dolinski K, Dwight SS, Engel S, Fisk DG, et al. Saccharomyces Genome Database (SGD) provides biochemical and structural information for budding yeast proteins. Nucleic Acids Res. 2003;31:216–218. doi: 10.1093/nar/gkg054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller LA, Zhang P, Rhee SY. AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132:453–460. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caspi R, Foerster H, Fulcher CA, Hopkinson R, Ingraham J, Kaipa P, Krummenacker M, Paley S, Pick J, et al. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2006;34:D1–D6. doi: 10.1093/nar/gkj128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green ML, Karp PD. The outcomes of pathway database computations depend on pathway ontology. Nucleic Acids Res. 2006;34:3687–3697. doi: 10.1093/nar/gkl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 10.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 11.Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, Garcia E, Halltorp G, Johansson D, et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- 12.Paley SM, Karp PD. Evaluation of computational metabolic-pathway predictions for H. pylori. Bioinformatics. 2002;18:715–724. doi: 10.1093/bioinformatics/18.5.715. [DOI] [PubMed] [Google Scholar]
- 13.Romero PR, Karp PD. Using functional and organizational information to improve genome-wide computational prediction of transcription units on pathway-genome databases. Bioinformatics. 2004;20:709–717. doi: 10.1093/bioinformatics/btg471. [DOI] [PubMed] [Google Scholar]
- 14.Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. The Comprehensive Microbial Resource. Nucleic Acids Res. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green ML, Karp PD. A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinformatics. 2004;5:76. doi: 10.1186/1471-2105-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]