Abstract
PharmGKB is a knowledge base that captures the relationships between drugs, diseases/phenotypes and genes involved in pharmacokinetics (PK) and pharmacodynamics (PD). This information includes literature annotations, primary data sets, PK and PD pathways, and expert-generated summaries of PK/PD relationships between drugs, diseases/phenotypes and genes. PharmGKB's website is designed to effectively disseminate knowledge to meet the needs of our users. PharmGKB currently has literature annotations documenting the relationship of over 500 drugs, 450 diseases and 600 variant genes. In order to meet the needs of whole genome studies, PharmGKB has added new functionalities, including browsing the variant display by chromosome and cytogenetic locations, allowing the user to view variants not located within a gene. We have developed new infrastructure for handling whole genome data, including increased methods for quality control and tools for comparison across other data sources, such as dbSNP, JSNP and HapMap data. PharmGKB has also added functionality to accept, store, display and query high throughput SNP array data. These changes allow us to capture more structured information on phenotypes for better cataloging and comparison of data. PharmGKB is available at www.pharmgkb.org
INTRODUCTION
The Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB; www.pharmgkb.org) is a public resource that promotes research into the relationships between human genotypes, phenotypes and clinical outcomes by linking and annotating primary data sets from ongoing research and established data from the literature (1,2). In addition to gene–drug relationships, the PharmGKB also contains data on gene variation, genomics, gene–disease relationships, drug action and pathways. The PharmGKB has developed highly curated pathways documenting the genes involved in pharmacodynamics and pharmacokinetics of a selection of drugs.
We have developed an XML format for defining genotype data, a relational database schema for data storage and a flexible mechanism for submitting phenotype data (3). We are also participating in the PML effort to define an XML standard for genotype/phenotype data exchange. PharmGKB first came online in April 2000, and major updates to the user interface appeared in 2002, 2004 and 2006. PharmGKB data and knowledge are updated on a continuous basis. Access is free but requires users to register for a username and password for viewing individual subject data.
DISTINGUISHING PHARMGKB USER GROUPS
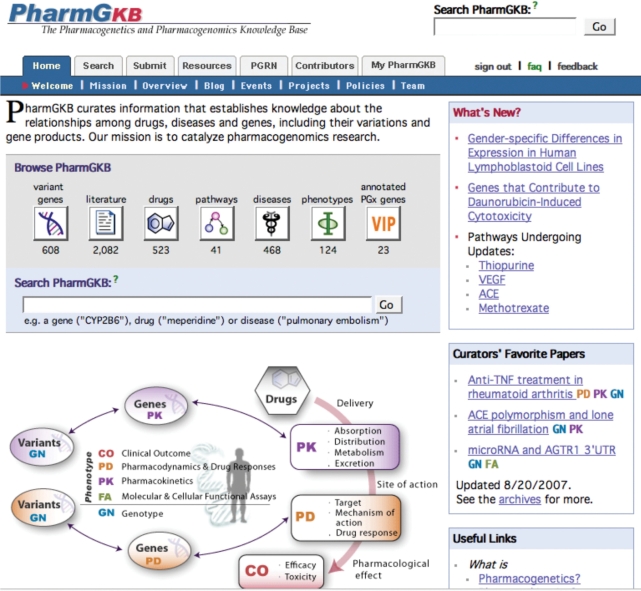
PharmGKB supplies a wide variety of pharmacogenomic knowledge to a broad range of users, from gene-based users to clinical scientists. In order to better serve our user groups, PharmGKB has redesigned its website homepage as well as the resource and submission tabs to more effectively disseminate knowledge in a form that matches user needs. PharmGKB has identified four main user groups of the database: gene-oriented users, drug-oriented users, bioinformaticians and clinical/disease-oriented investigators. PharmGKB organizes knowledge pertinent to each group into user-based views and resources. Figure 1 depicts the PharmGKB homepage that provides direct links to different user-based knowledge, such as drug pages or gene pages and more specifically pharmacokinetic and pharmacodynamic data.
Figure 1.
PharmGKB disseminates highly curated pharmacogenomic knowledge in a form that matches user needs.
INTEGRATING GENOME-SCALE DATA
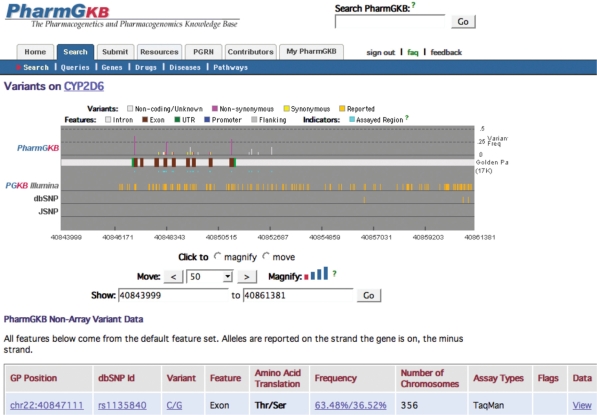
High-throughput functional genomic technologies have resulted in the rapid accumulation of genome-scale data sets. There is a renewed emphasis on genetic variation, partly due to the haplotype mapping project undertaken by HapMap(4). The interest lies in the notion that a single nucleotide polymorphism (SNP) can contribute directly to disease predisposition by modifying a gene's function, or that SNPs can be used as genetic markers to tag near neighbor disease causing mutations through association studies and linkage analysis. Genetic variation is now measured on a genomic scale using SNP arrays. The successful analysis of such data sets depends on rapid access to the most current annotation of the SNPs being studied in conjunction with phenotypic data, linkage disequilibrium information and other genomic data (5). Accordingly, PharmGKB has added functionality to integrate, aggregate and annotate data from genome-wide studies. These data can be queried and viewed by chromosome browsing, gene pages or individual submission pages. Figure 2 shows the ABCB1 gene page that has been populated with an additional 15 variants from an Illumina 317K SNP array, assayed both from traditional and high-throughput methods.
Figure 2.
PharmGKB VariantPage. CYP2D6 variant page contains information on the variants within the gene boundaries. SNP array data are integrated with traditional PharmGKB primary data and other external variant data, such as dbSNP and JSNP.
VIP GENE AND VARIANT PAGES
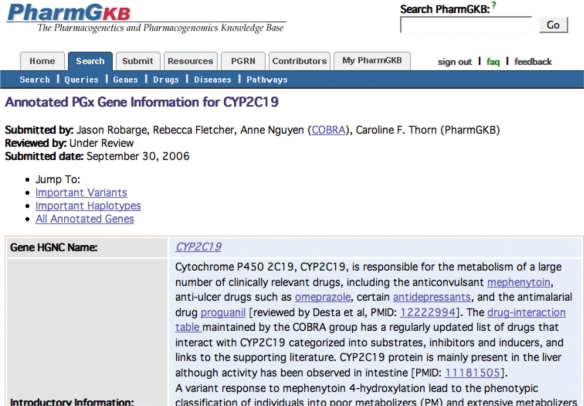
Very Important Pharmacogenes, provide annotated information about genes, variants, haplotypes and splice variants of particular relevance for pharmacogenetics and pharmacogenomics. VIP gene pages highlight the key variants, haplotypes, drugs, diseases and phenotypes associated with the pharmacogene (Figure 3). A VIP gene is defined as a gene that has well-documented information about its involvement in the pharmacodynamics or pharmacokinetics of a drug. There are a total of about 200 well-documented VIP genes that were selected by pharmacogenomics experts for PharmGKB to annotate. VIP pages are hand-curated and contain information about the mapping of the variant to allow cross comparison with other resources, frequencies and drug and disease associations from key studies and the links to the literature that document them. VIP variants link from the variant page via a flag in the row for that golden path position.
Figure 3.
PharmGKB very important pharmacogenomic (VIP) genes: PharmGKB gathers knowledge on key pharmacogenomic genes, which highlight important variants, haplotypes, drugs, diseases and phenotypes associated with the gene.
PHARMGKB PATHWAYS
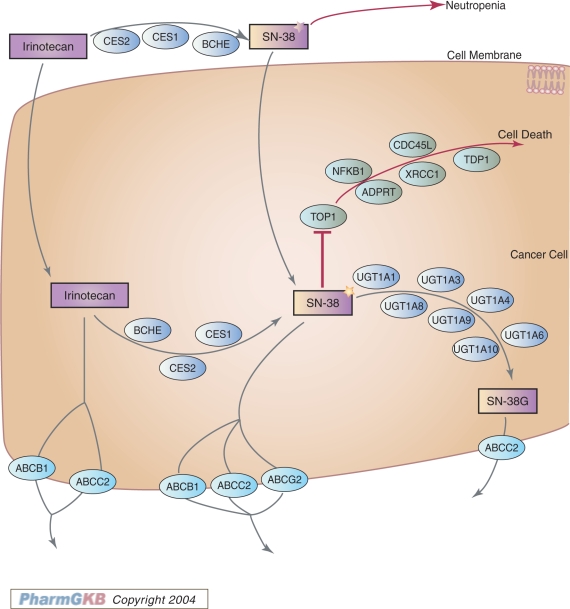
Historically, many pharmacogenetic studies have focused on single genes involved in drug side-effects. There is now a growing interest in how pathways of interacting genes can affect both drug metabolism and drug response. PharmGKB pathways are drug-centered, depicting candidate genes for pharmacogenetics and pharmacogenomics studies and they provide the means to connect separate data sets to represent the current knowledge as a cohesive snapshot. Pathways are created based on community interest and involvement/contributions. The diagrams have information content in the shape and color of the icons that represent whether the component is a gene, drug, metabolic intermediate, etc. (Figure 4). The pathways are interactive: clicking on a gene takes the user to the gene page from which available genotype and phenotype data and literature citations can be found. Drugs and metabolites are represented by rectangles. Clicking on a drug takes the user to the drug page from which available phenotype data and literature citations can be found. Clicking on a golden arrow presents the user with phenotype data that support a relationship. In addition to the pathway diagram, a summary is provided to describe the content of the graphic. The pathways are generated by collaboration of investigators and represent a consensus of the opinions of the authors. Currently, these pathways are constructed by hand as graphic images and updated by the scientific community every 2 years. Dates of pathway release and updates are posted on the pathway pages.
Figure 4.
PharmGKB Irinotecan Pathway. View of a model human liver cell showing blood, bile, and intestinal compartments, indicating tissue-specific involvement of genes in the irinotecan pathway. Drugs are depicted by purple boxes, transporter genes by turquoise ovals and genes coding for metabolic enzymes by blue ovals. Available at: http://www.pharmgkb.org/search/pathway/irinotecan/liver.jsp.
LITERATURE ANNOTATIONS
PharmGKB has a collection of pharmaco-related literature annotations that are generated and enhanced through hand-curation. We allow reference addition to PharmGKB either as a Publication entry or as a Literature Annotation, which includes additional hand-curated details about the reference and the pharmacogenomic relationships described in the article. To aid users, PharmGKB knowledge generation is achieved by combining highly searchable controlled vocabulary classifications of references with brief, detailed free-text descriptions of the primary research findings. This provides users with the most flexible access to the knowledge we generate about the references, depending on their different needs and preferences.
PHENOTYPE DATA SETS
PharmGKB houses a variety of phenotype data sets. All phenotype submissions are accepted and PharmGKB has expanded submission methods to include Microsoft Office documents, or alternatively, a URL to another established archival public database (e.g. GEO). High-impact phenotype data sets are curated by hand, while others receive minimal oversight at PharmGKB. High-impact phenotype data sets correspond to genotype data submitted to PharmGKB and are typically published in peer-review journals. These data are featured on the PharmGKB website with an interactive display, curated annotations and downloadable Excel files. All phenotype files on PharmGKB are fully text-searchable. PharmGKB also offers links to web sites with controlled vocabularies in order to encourage investigators to optimize documentation of their deposits.
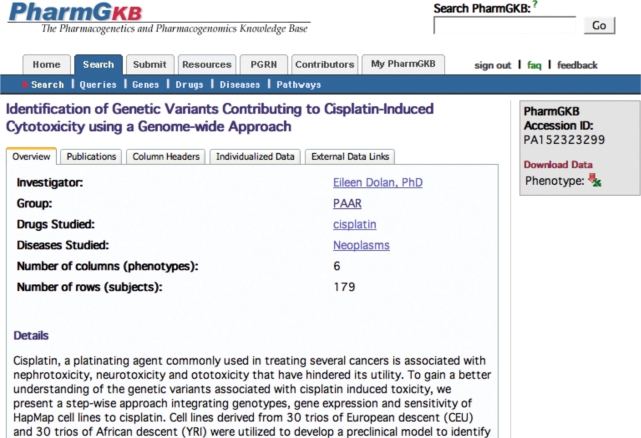
The phenotype data set knowledge is available to the general public via an integrated tab-display (Figure 5). For all data sets in PharmGKB, genes, drugs, diseases and categories of pharmacogenetic evidence are tagged and indexed for querying. Curated phenotype data sets are reviewed and include meaningful phenotypic annotations related to pharmacogenomic research, from clinical-metabolite data to protein constructs. Genotype and/or phenotype data can be downloaded by clicking on arrows in the upper right corner of the webpage.
Figure 5.
PharmGKB phenotype data set. PharmGKB displays phenotype data. In the figure, there are five tabs: (i) Overview presents key indexing terms and a summary of the file, (ii) Publications points to key publication summaries, (iii) Column Headers defines the data present in the file and (iv) Individualized Data provides a data browser to look at the primary data. (v) External Data Links point to external data sources relevant to the data set, such as exon arrays stored in other databases.
FUTURE DIRECTIONS
PharmGKB catalyzes the generation of new knowledge in pharmacogenomics through the development, implementation and dissemination of a public resource focused on both data and knowledge. In the short-term, this resource will facilitate basic research. In the long-term, it will impact how medicine is delivered. Our future work focuses on detailed annotation of individual human polymorphisms (or haplotypes) that are important for drug response phenotypes. We also are active in creating consortia of investigators interested in pooling pharmacogenomic data sets in order to improve population coverage and statistical power.
ACKNOWLEDGEMENTS
PharmGKB is supported by NIH/NIGMS Grant no. UO1GM61374. Funding to pay the Open Access publication charges for this article was provided by NIH/NIGMS grant no. UO1GM61374.
Conflict of interest statement. None declared.
REFERENCES
- 1.Thorn CF, Klein TE, Altman RB. PharmGKB: the pharmacogenetics and pharmacogenomics knowledge base. Methods Mol. Biol. 2005;311:179–191. doi: 10.1385/1-59259-957-5:179. [DOI] [PubMed] [Google Scholar]
- 2.Hewett M, Oliver DE, Rubin DL, Easton KL, Stuart JM, Altman RB, Klein TE. PharmGKB: the pharmacogenetics knowledge base. Nucleic Acids Res. 2002;30:163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whirl-Carrillo MMW, Thorn CF, Klein TE, Altman RB. The PharmGKB XML Schema: an interchange format for genotype-phenotype databases. Human Mutation. 2007 doi: 10.1002/humu.20662. In press. [DOI] [PubMed] [Google Scholar]
- 4.Consortium IH. A haplotype map of the Human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernandez-Boussard T, Klein TE, Altman RB. Pharmacogenomics: the relevance of emerging genotyping technologies. MLO Med. Lab. Obs. 2006;38, 24:26–30. [PubMed] [Google Scholar]