Abstract
The Functional Single Nucleotide Polymorphism (F-SNP) database integrates information obtained from 16 bioinformatics tools and databases about the functional effects of SNPs. These effects are predicted and indicated at the splicing, transcriptional, translational and post-translational level. As such, the database helps identify and focus on SNPs with potential deleterious effect to human health. In particular, users can retrieve SNPs that disrupt genomic regions known to be functional, including splice sites and transcriptional regulatory regions. Users can also identify non-synonymous SNPs that may have deleterious effects on protein structure or function, interfere with protein translation or impede post-translational modification. A web interface enables easy navigation for obtaining information through multiple starting points and exploration routes (e.g. starting from SNP identifier, genomic region, gene or target disease). The F-SNP database is available at http://compbio.cs.queensu.ca/F-SNP/.
INTRODUCTION
Much effort in current human genomics, epidemiology and pharmacogenomics is focused on the identification of genetic variations that are responsible for common and complex diseases. Specifically, single nucleotide polymorphisms (SNPs), which are substitutions of a single nucleotide at a specific position on the genome, are in the forefront of such studies, as they form the majority of genetic variations in the human population. Reliable identification of disease-causing SNPs is expected to enable early diagnosis, personalized treatment and targeted drug design.
The F-SNP database gathers computationally predicted functional information about SNPs, particularly aiming to facilitate identification of disease-causing SNPs in association studies. Due to the large overhead of large-scale genotyping and analysis, it is often required, when conducting association studies, to prioritize SNPs in a target genomic region based on their potential functional effects (1). Typically, SNPs occurring in functional genomic regions such as protein coding or regulatory regions are more likely to cause functional distortion and, as such, more likely to underlie disease-causing variations. Current bioinformatics tools examine the functional effects of SNPs only with respect to a single biological function. Therefore, much time and effort is required from researchers to separately use multiple tools and interpret the (often conflicting) predictions.
To help expedite the process, the F-SNP database aims to provide a comprehensive collection of functional information about SNPs, using a large variety of publicly available tools and resources. Specifically, it provides information about potential deleterious effects of SNPs with respect to four major biomolecular functional categories, namely, splicing, transcription, translation and post-translation. Moreover, for assessing the deleterious effect of SNPs along each functional category, F-SNP integrates multiple tools that are based on different algorithms, data and resources. No single tool can yet capture all the possible effects of SNPs on even one biological function (2). Providing predictions from multiple diverse methods thus helps to better assess the functional impact of each SNP. Researchers can also use the raw predictions provided by F-SNP to implement their own tool for evaluating functional effects of SNPs.
Another distinguishing feature of the F-SNP database is its integration of human-disease databases to facilitate identification of potential disease-causing SNPs as genetic markers in association studies. The F-SNP database provides a web interface that takes as input either a disease, a gene, a genomic region or a SNP identifier. If the input is a specific disease, its candidate genes, obtained from the integrated human-disease databases, are provided with their SNP information. Thus, researchers interested in a specific disease can retrieve a list of all the candidate genes relevant to this disease along with functional information for all the SNPs within each candidate gene as predicted by a variety of bioinformatics tools.
The current version of the F-SNP database contains the functional information for 559 322 SNPs in 18 282 genes relevant to 85 major human diseases. Currently, functional assessment of SNPs is done by 16 bioinformatics tools and databases. The following sections describe the procedure used for constructing the F-SNP database, provide a brief description of its current contents, and explain the web-based interface.
DATABASE CONSTRUCTION
SNPs and genes
We downloaded the dataset of 11 811 594 human SNPs and their annotations from the dbSNP (build 126) (3) and Ensembl (release 42) (4) databases. We also downloaded a list of 38 550 human genes along with their primary information such as gene symbol, alias names, chromosomal location and gene type from NCBI Entrez Gene (downloaded 12 December 2006).
SNP to gene mapping
To link SNPs with specific genes, for each gene, SNPs located along the gene region (including 5 kb upstream and 5 kb downstream) were identified. A total of 4 043 147 SNPs are thus mapped to 23 630 human genes.
Gene to disease mapping
We retrieved from NCBI's Genes and Disease site the list of 85 human genetic disorders, categorized by the 16 body parts that they affect (downloaded 29 January 2007). To link candidate genes with the 85 diseases, we downloaded the dataset of a gene-disease map from NCBI's OMIM database (downloaded 3 January 2007) (5). Accordingly, 2374 genes were mapped to 85 human genetic disorders.
Assessing the functional effects of SNP
Using a variety of publicly available bioinformatics tools, we assess the functional effects of SNPs along the following four major categories: protein coding, splicing regulation, transcriptional regulation and post-translation effects. The tools, PolyPhen (as of 15 August 2007) (6), SIFT (as of 15 August 2007) (7), SNPeffect (version 2.0) (8), SNPs3D (as of 15 August 2007) (9) and LS-SNP (as of 15 August 2007) (10) are used to identify non-synonymous deleterious SNPs; ESEfinder (release 3.0) (11), RescueESE (as of 15 August 2007) (12), ESRSearch (as of 15 August 2007) (13) and PESX (as of 15 August 2007) (14) are used to identify SNPs in exonic splice regions; The Ensembl database (release 42) (4) is used to identify nonsense SNPs and SNPs in intronic splice sites; TFSearch (ver. 1.3) (15) and Consite (as of 15 August 2007) (16) are used to identify transcriptional regulatory SNPs in promoter regions; The Ensembl (release 42) (4) and GoldenPath (downloaded 12 December 2006) (17) databases are used to identify SNPs in other transcriptional regulatory regions (e.g. microRNA, cpgIslands); KinasePhos (as of 15 August 2007) (18), OGPET (ver. 1.0) (19) and Sulfinator (as of 15 August 2007) (20) are used to examine post-translation modification sites. In addition, genomic regions that are conserved across multiple species are identified using GoldenPath (downloaded 12 December 2006) (17), and are used as described below. The complete list of 16 integrated tools and databases is provided in Table 1.
Table 1.
Bioinformatics tools and databases integrated into F-SNP (Release 1.0. August 2007)
For each possible functional category into which a SNP may be classified, the table provides the tools that examine this function, and the URL from which the respective tool is available (as of August 2007). The category Conserved Region in the last row is not a functional category in-and-of itself, but is informative in determining the effect of SNPs on splicing and transcriptional regulation.
Summarizing the functional importance of SNPs
In addition to providing the raw output from the 16 integrated tools stating the functional effects of SNPs, F-SNP also denotes a subset of the assessed SNPs as ‘functional’ SNPs; these are SNPs that are predicted by a majority of the integrated tools to be deleterious with respect to at least one biological function of a gene or a gene product.
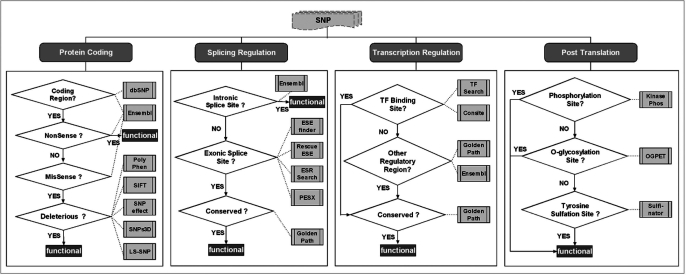
Figure 1 illustrates the assessment process. We note that in the case of SNPs within regulatory regions, for instance, ‘transcription factor binding site’ or ‘exonic splicing regulatory regions’ (as shown in the two middle boxes in Figure 1), we additionally examine whether the region is conserved across multiple species (chimp/dog/mouse/rat/chicken/zebrafish/fugu) to determine whether the SNP is functional. This strategy is mainly used because there is a high rate of false positive findings by in silico prediction tools due to the short length of such sequences (typically 6–8-mer) (12). The additional information about conserved regions across multiple species is thus used as a way to filter out possible false-positive predictions (2,11–14).
Figure 1.
Decision procedure for functional SNP assessment. Each SNP is examined for deleterious effects with respect to each functional category (i.e. protein coding, splicing regulation, transcriptional regulation and post-translation—as shown in the top part of the figure). For each category, a series of tests is executed to determine whether the SNP has a functional impact. First the type (coding, intronic, etc.) of the genomic region is identified, using data from dbSNP (3) and Ensembl (4). Once this is determined, other tests are performed. For example, to assess if a SNP has a deleterious effect on protein coding, it first must be located on a coding region. Ensembl (4) is used to examine if this is a nonsense mutation, in which case the SNP is considered to be deleterious. Otherwise—if the SNP is a missense mutation, it is further tested by five different tools [PolyPhen (6), SIFT (7), SNPeffect (8), SNPs3D (9) and LS-SNP (10)] to check if the non-synonymous substitution is deleterious. A majority vote among these tools concludes the process, and identifies the SNP as either having a potentially deleterious functional impact (denoted ‘functional’ in the figure) or not.
DATABASE CONTENTS
The F-SNP database, release 1.0 (August 2007), contains the assessed functional information for 559 322 SNPs within 18 282 candidate genes for 85 human diseases. Detailed statistics of the current F-SNP database are provided in Table 2. The database will be continuously updated to provide functional information about additional SNPs.
Table 2.
Statistics of functionally assessed SNPs in F-SNP, Release 1.0 (August 2007)
| Functional category | Number of assessed SNPs | Number of potentially deleterious SNPs |
|---|---|---|
| Protein coding | 154 140 | 66 899 |
| Splicing regulation | 73 051 | 8 075 |
| Transcriptional Regulation | 453 710 | 78 296 |
| Post-translation | 64 736 | 4 477 |
| Total | 559 322 | 115 356 |
For each functional category, the number of SNPs for which the function has been assessed using the 16 tools and databases integrated into F-SNP is shown in the middle column. The number of SNPs indicated by F-SNP to be potentially deleterious is shown on the right.
WEB INTERFACE
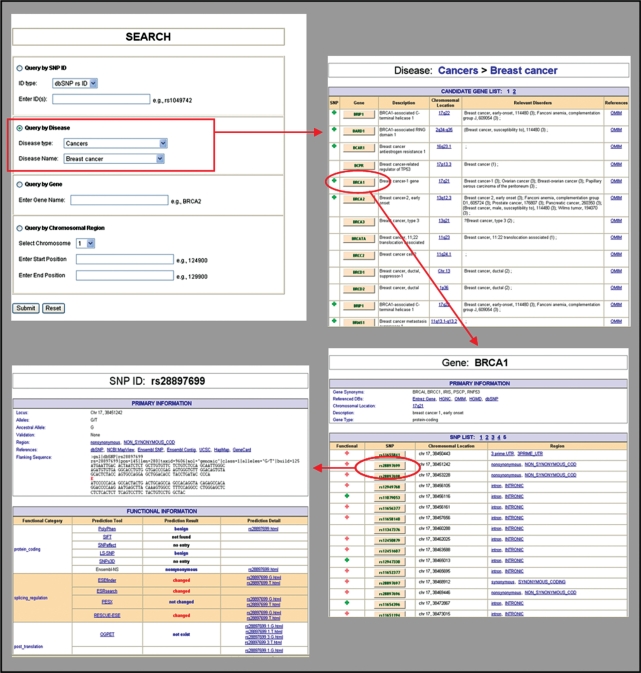
The F-SNP database is available at http://compbio.cs.queensu.ca/F-SNP/. The user can search the database by SNP identifier, gene, disease or chromosomal regions. Figure 2 shows an example of results obtained from an interactive search concerned with breast cancer.
Figure 2.
Example of an F-SNP search session. (a) The initial search page is displayed, where the user selected the disease type to be Cancers, and the specific disease to be Breast cancer (Search by disease). (b) Results obtained after clicking the Submit button in panel (a), namely a list of genes associated with Breast cancer along with their associated chromosome location, known related disorders, and links to OMIM. The BRCA1 link (circled) is selected and clicked. (c) A detailed description of SNPs associated with BRCA1 is produced (demonstrating results of Search by gene). The SNP whose identifier is rs28897699 (circled)—indicated by a ‘+’ mark to have associated functional information—is selected and clicked. (d) Information about the SNP rs28897699 is presented (demonstrating results of Search by SNP ID).
Search by SNP identifier
To obtain information about a single SNP the database can be searched by providing the SNP's rs-identifier from dbSNP (build 126) (3). The resulting page provides the primary information about the SNP along with its assessed functional information. The primary information includes the chromosomal location of the SNP, alleles, ancestral allele, validation status, type of genomic region, links to external databases namely dbSNP (build 126) (3), NCBI MapView (homo sapiens build 125), Ensembl (release 42) (4), Ensembl Contig (as of 15 August 2007), UCSC Genome Browser (March 2006 assembly) (17), HapMap (Rel 21a/phase II) (21) and GeneCards (ver. 2.37) (22), and the flanking sequence around the SNP. The functional information provided for each SNP includes functional category, integrated tools used, prediction results and the detailed output from each predictive tool.
Search by gene
To find the SNPs located within a specific gene region, the database can be searched by providing the HUGO name of the gene or of its protein. If no official HUGO name matches the input keyword, alias gene names (registered in NCBI Entrez Gene) are examined for the search. A table with all the SNPs linked to the gene is then produced, where a green ‘+’ mark is shown next to each SNP for which the functional effects have been assessed, and a red ‘+’ mark further indicates that the SNP was determined to have a potentially deleterious functional effect. The user can then click on each SNP to obtain the detailed functional information about it.
Search by disease
To identify SNPs that may be related to a specific disease the user can select the disease category and name. A table with all the genes relevant to the disease is produced. The user can then click on each gene to go to the gene-information page. As described earlier, the gene-information page lists all the SNPs linked to the gene, for which the user can retrieve further information.
Search by chromosomal region
To study SNPs along a chromosomal region the user can provide the chromosome number, along with start/end positions. A table with all the SNPs within the region is produced and, as explained earlier, a ‘+’ mark indicates the SNPs for which functional effects have been assessed. Again, the user can click on each SNP to obtain further information.
CONCLUSIONS AND FUTURE WORK
The F-SNP database is a comprehensive resource collecting computationally obtained functional information about SNPs. The information is given in four levels, namely, protein coding, splicing regulation, transcriptional regulation and post-translation. As effective association studies largely depend on prioritizing the SNPs to be examined and studied, we expect that F-SNP will serve as a one-stop tool for selecting potential disease-causing SNP markers for association studies. The functional information provided for SNPs will be regularly updated as other prediction tools and biomolecular experiments become available. We also plan to integrate additional human-disease databases to include a broader spectrum of common and complex diseases.
ACKNOWLEDGEMENTS
This work is supported by HS's NSERC Discovery Grant 298292-04 and CFI New Opportunities Award 10437, and by PL's Ontario Graduate Scholarship and Duncan & Urlla Carmichael Graduate Fellowship. The Open Access publication charges were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brunham LR, Singaraja RR, Pape TD, Kejariwai A, Thomas PD, Hayden MR. Accurate prediction of the functional significance of single nucleotide polymorphisms and mutations in the ABCA1 gene. PLoS Genet. 2005;1:739–747. doi: 10.1371/journal.pgen.0010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatti P, Church D, Rutter JL, Struewing JP, Sigurdson AJ. Candidate single nucleotide polymorphism selection using publicly available tools: a guide for epidemiologists. Am. J. Epidemiol. 2006;164:794–804. doi: 10.1093/aje/kwj269. [DOI] [PubMed] [Google Scholar]
- 3.Sherry S, Ward M, Kholodov M, Baker J, Phan L, Smigielski E, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkl996. (Database issue), d1-d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKusick-Nathans Institute of Genetic Medicine. John's Hopkins University and National Center for Biotechnology Information, NLM. Online Mendelian Inheritance in Man, OMIM ™. http://www.ncbi.nlm.nih.gov/omim/.
- 6.Ramensky V, Sunyaev S. Human nonsynonymous SNPs: server and survey. Nucleic Acid Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng P, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reumers J, Schymkowitz J, Ferkinghoff-Borg J, Stricher F, Serrano L, Rousseau F. SNPeffect: a database mapping molecular phenotypic effects of human non-synonymous coding SNPs. Nucleic Acid Res. 2005;33(Database issue):D527–D532. doi: 10.1093/nar/gki086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karchin R, Diekhans M, Kelly L, Thomas D, Pieper U, Eswar N, Haussler D. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 11.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeo G, Burge CB. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl Acad. Sci. USA. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 14.Zhang XH-F, Kangsamaksin T, Chao MSP, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol. Cell. Biol. 2005;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akiyama Y. 1998. TFSEARCH: searching transcription factor binding sites. http://www.rwcp.or.jp/papia/. [Google Scholar]
- 16.Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32(Web Server issue):W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn R, Karolchik D, Zweig A, Trumbower H, Thomas D, Thakkapallayil A, Sugnet C, Stanke M, Smith K, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35(Database issue):D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang H, Lee T, Tseng S, Horng J. KinasePhos: a web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res. 2005;33(Web server issue):W226–229. doi: 10.1093/nar/gki471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerken T, Tep C, Rarick J. The role of peptide sequence and neighboring residue glycosylation on the substrate specificity of the uridine 5′-diphosphate-alpha-n-acetylgalactosamine:polypeptide n-acetylgalactosaminyl transferases t1 and t2: kinetic modeling of the porcine and canine submaxillary gland mucin tandem repeats. Biochemistry. 2004;43:9888–9900. doi: 10.1021/bi049178e. [DOI] [PubMed] [Google Scholar]
- 20.Monigatti F, Gasteiger E, Bairoch A, Jung E. The Sulfinator: predicting tyrosine sulfation sites in protein sequences. Bioinformatics. 2002;18:769–770. doi: 10.1093/bioinformatics/18.5.769. [DOI] [PubMed] [Google Scholar]
- 21.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 22.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. Genecards: encyclopedia for genes, proteins and diseases. 1997. http://www.genecards.org/Weizmann Institute of Science, Bioinformatics Unit and Genome Center, Israel. [DOI] [PubMed]