Abstract
KEGG (http://www.genome.jp/kegg/) is a database of biological systems that integrates genomic, chemical and systemic functional information. KEGG provides a reference knowledge base for linking genomes to life through the process of PATHWAY mapping, which is to map, for example, a genomic or transcriptomic content of genes to KEGG reference pathways to infer systemic behaviors of the cell or the organism. In addition, KEGG provides a reference knowledge base for linking genomes to the environment, such as for the analysis of drug-target relationships, through the process of BRITE mapping. KEGG BRITE is an ontology database representing functional hierarchies of various biological objects, including molecules, cells, organisms, diseases and drugs, as well as relationships among them. KEGG PATHWAY is now supplemented with a new global map of metabolic pathways, which is essentially a combined map of about 120 existing pathway maps. In addition, smaller pathway modules are defined and stored in KEGG MODULE that also contains other functional units and complexes. The KEGG resource is being expanded to suit the needs for practical applications. KEGG DRUG contains all approved drugs in the US and Japan, and KEGG DISEASE is a new database linking disease genes, pathways, drugs and diagnostic markers.
INTRODUCTION
Since the completion of the Human Genome Project, high-throughput experimental projects have been initiated for uncovering genomic information in an extended sense, including transcriptome and proteome, as well as metabolome, glycome and other genome-encoded information. Together with traditional genome sequencing for an increasing number of organisms, we are beginning to understand the genomic space of possible genes and proteins that make up the biological system. In contrast, we have very limited knowledge about the chemical space of possible chemical substances that exists as an interface between the biological world and the natural world. This situation is rapidly changing thanks to the chemical genomics initiatives for systematic screening of biologically active chemical compounds and the metagenomics initiatives giving insights into the chemical environment that interacts with and drives evolution of the biological system.
The KEGG project was initiated in 1995, coincidentally when the first genome of a free-living organism was completely sequenced (1). KEGG PATHWAY has since been utilized as a reference knowledge base for understanding higher-level functions of cellular processes and organism behaviors from large-scale molecular data sets. The addition of KEGG BRITE, a collection of functional hierarchies with structured vocabularies, significantly increased our ability to represent and utilize higher-level functional information, especially to integrate genomic and chemical (environmental) information (2). Here we report another new development in KEGG, the integration of research results and practical values in medical, pharmaceutical and environmental sciences.
THE KEGG RESOURCE
Overview
As of January 2008, KEGG comprises 19 databases, categorized into systems information, genomic information and chemical information as shown in Table 1. The six databases in the chemical information category are collectively called KEGG LIGAND. The six databases in the lower part of the genomic information category are computationally generated, but all the other 13 databases are manually curated.
Table 1.
KEGG databases
| Category | Database | Content |
|---|---|---|
| Systems information | KEGG PATHWAY | Pathway maps |
| KEGG BRITE | Functional hierarchies | |
| KEGG MODULE | Pathway modules (released January 2008) | |
| KEGG DISEASE | Diseases (released January 2008) | |
| Genomic information | KEGG ORTHOLOGY | KEGG orthology (KO) groups |
| KEGG GENOME | KEGG organisms | |
| KEGG GENES | Genes in high-quality genomes | |
| KEGG DGENES | Genes in draft genomes | |
| KEGG EGENES | Genes as EST contigs | |
| KEGG VGENOME | Viral genomes (to be fully integrated) | |
| KEGG VGENES | Genes in viral genomes (to be fully integrated) | |
| KEGG OGENES | Genes in organelle genomes (to be fully integrated) | |
| KEGG SSDB | Sequence similarities and best hit relations | |
| Chemical information | KEGG COMPOUND | Metabolites and other chemical compounds |
| KEGG DRUG | Drugs | |
| KEGG GLYCAN | Glycans | |
| KEGG ENZYME | Enzymes | |
| KEGG REACTION | Enzymatic reactions | |
| KEGG RPAIR | Reactant pairs and chemical transformations |
The KEGG databases are highly integrated. In fact, KEGG should be viewed as a computer representation of the biological system, where biological objects and their relationships at the molecular, cellular and organism levels are computerized as separate database entries. Each database entry, called a KEGG object, is given a unique identifier within KEGG. Table 2 summarizes the naming convention of such KEGG object identifiers for the 13 core databases. Except for GENES and ENZYME that utilize the standard names of locus_tag and EC number, and for GENOME that distinguishes organisms with 3–4 letter KEGG organism codes, the KEGG object identifier is a five-digit number prefixed by an upper-case alphabet or a 2–4 letter code (map, br or organism code). Examples are: C00047 for lysine, K04527 for insulin receptor and hsa05210 for colorectal cancer pathway.
Table 2.
KEGG object identifiers
| Release | Database | Object identifier |
|---|---|---|
| 1995 | KEGG PATHWAY | map number |
| KEGG GENOME | organism code (T number) | |
| KEGG GENES | locus_tag/NCBI GeneID | |
| KEGG ENZYME | EC number | |
| KEGG COMPOUND | C number | |
| 2001 | KEGG REACTION | R number |
| 2002 | KEGG ORTHOLOGY | K number |
| 2003 | KEGG GLYCAN | G number |
| 2004 | KEGG RPAIR | A number |
| 2005 | KEGG BRITE | br number |
| KEGG DRUG | D number | |
| 2008 | KEGG MODULE | M number |
| KEGG DISEASE | H number |
See http://www.genome.jp/kegg/kegg3.html for details.
These identifiers may be used to directly obtain corresponding database entries with the ‘Get Entry’ option in the KEGG website (http://www.genome.jp/kegg/). Interestingly, these identifiers may also be used in web search engines, such as Google and Yahoo, to obtain corresponding KEGG database entries. There are already many databases that are linked to/from KEGG. Such outside links will continue to be added to better integrate KEGG with various other web resources.
Genome annotation
Genome annotation in KEGG assigns KO (KEGG Orthology) identifiers or K numbers to genes in a single genome or simultaneously to genes in multiple genomes. With the addition or revision of a KEGG pathway map or BRITE hierarchy, KO groups (K numbers) are defined for the pathway nodes (boxes) or the hierarchy nodes (bottom leaves). Then the corresponding genes in selected organisms (usually in the literature) are manually annotated with the new K numbers, which are reflected in KEGG GENES. Thus, KEGG GENES can be used as a reference database for genome annotation. The number of KO groups has been increasing at a rate of about 2000 per year, and it is now over 10 000.
The KO assignment is applied to a new genome as follows. First, the new genome is subject to SSDB computation, a comparison of protein coding genes against all existing genomes by the SSEARCH program. The result is stored in KEGG SSDB containing sequence similarity scores and best-hit information for all gene pairs. Then, computational KO assignment is done by the KAAS-SSDB program, followed by manual verification and additional assignment with the GFIT tool. An automated version of this genome annotation procedure is made available as the KAAS web service (3), which utilizes BLAST rather than SSEARCH for pairwise genome comparisons.
The KO system is the basis for linking genomes to biological systems through the process of pathway mapping and BRITE mapping. For each organism in KEGG, organism-specific pathways and BRITE hierarchies are computationally generated based on its assigned K numbers. Microarray gene expression profile data may then be mapped to these pathways and hierarchies to infer systemic functions of the cell or the organism. In addition to the hierarchies of genes and proteins (K numbers), KEGG BRITE contains the hierarchies of chemical substances (C, D, G, R numbers) together with known relationships to K numbers, such as ligand–receptor interactions and drug–target relationships. By using these relationships, the BRITE mapping will be improved to present clues for understanding the interactions with the environments.
Chemical annotation
The KO system can also be used for chemical annotation, which is the linking of genomic or transcriptomic contents of genes to chemical structures of endogenous molecules. This is achieved by finer classifications of KO groups for specific classes of enzymes distinguishing different substrate specificity, as well as accumulating knowledge of biosynthetic pathways. For example, glycans are synthesized by a series of reactions catalyzed by glycosyltransferases. With the KEGG pathway maps for glycan structures (map01030 and map01031) or the KEGG GLYCAN composite structure map (4), where edges (glycosidic linkages) correspond to K numbers (glycosyltransferase orthologs), the gene content in the genome can be converted to possible glycan structures. In a similar but more sophisticated way, glycan structures can be predicted from microarray gene expression data (5). The KEGG resource will be made suitable to cope with the diversity of other molecules as well, including polyketides/non-ribosomal peptides (6), polyunsaturated fatty acids and terpenoids.
Another type of chemical annotation is to characterize biological meaning in the chemical structures of small molecules. As reported previously (2), the knowledge of enzymatic reactions and associated chemical structure transformations is stored in KEGG REACTION and KEGG RPAIR. Each structure transformation is characterized by the RDM pattern (7), and most of the patterns are found uniquely or preferentially in specific categories of KEGG pathways (8). This tendency was used to predict the metabolic fate of xenobiotic chemical compounds. Software for reaction/pathway prediction is being developed as an upgrade of e-zyme and PathComp in KEGG LIGAND.
Enhancements to KEGG pathway
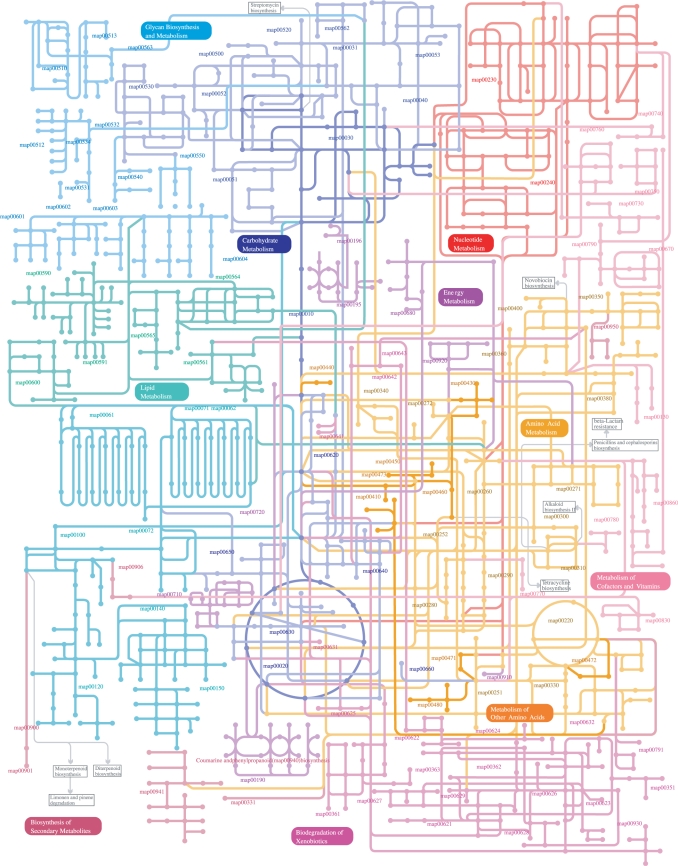
KEGG PATHWAY has been significantly expanded over the last 2 years with the addition of about 50 new pathway maps, mostly for signal transduction, cellular processes and human diseases. However, the traditional KEGG metabolic pathway maps are still most widely used including the KGML (KEGG XML) version. They are now supplemented with two new features introduced as a response to user feedback. The first feature is a global map shown in Figure 1, which is created as an SVG file by manually combining about 120 existing maps. Each node (circle) is a chemical compound and each line (curved or straight) connecting two nodes is a series of reactions (one to several reactions), which is also manually defined as a segment lacking branches. The new KEGG metabolism map allows the user to view and compare the entire metabolism, such as by mapping metagenomics data or microarray data. KGML users should also find the new KEGG metabolism map much easier to manipulate.
Figure 1.
The new KEGG metabolism map created as an SVG file.
The other feature is KEGG MODULE, a new database that collects pathway modules and other functional units as a set of K numbers. Pathway modules are smaller pieces of subpathways (see the BRITE hierarchy ko00002), manually defined as consecutive reaction steps, operon or other regulatory units, phylogenetic units obtained by genome comparisons, etc. This new database also contains molecular complexes, facilitating better organization of data and knowledge, especially in KEGG BRITE. The hierarchy of molecular organization, such as the subunit organization of transporters or receptors, is represented by the M number that corresponds to a set of K numbers. Incidentally, a line segment in the new KEGG metabolism map that also corresponds to a set of K numbers is identified by the N number, representing a mechanistically defined network segment.
KEGG for medical and pharmaceutical applications
As of September 2007, KEGG PATHWAY contains 26 maps for human diseases, among which 19 were introduced in the last 2 years. The disease pathway maps are classed in four subcategories: 6 as neurodegenerative disorders (9), 3 as each of infectious diseases and metabolic disorders and 14 as cancers. Although such maps will continue to be added, they will never be sufficient to represent our knowledge of molecular mechanisms of diseases because in many cases it is too fragmentary to represent as pathways. KEGG DISEASE is another addition to the KEGG suite of databases accumulating molecular-level knowledge on diseases including genes, drugs and biomarkers. Our current effort is focused on the four subcategories of diseases mentioned above.
The number of entries in KEGG DRUG has also significantly increased over the last 2 years, and now covers all approved drugs in the US and Japan. KEGG DRUG is a structure-based database. Each entry is a unique chemical structure that is linked to standard generic names, and is associated with efficacy and target information as well as drug classifications. Target information is presented in the context of KEGG pathways and drug classifications are part of KEGG BRITE. The generic names are linked to trade names and subsequently to outside resources of package insert information (patient information) whenever available. This reflects our effort to make KEGG more useful to the general public.
ACCESSING KEGG
Via GenomeNet
KEGG is made available as the major component of the Japanese GenomeNet service, operated by the Kyoto University Bioinformatics Center. The top pages of the KEGG website (http://www.genome.jp/kegg/) have been changed for easier access to KGML, KEGG API and KEGG FTP.
Via the new site
Because the KEGG system has become so large and complex, the entire package is being redesigned and is presented at a new site (http://www.kegg.jp/) that currently contains a Japanese version only.
ACKNOWLEDGEMENTS
THE KEGG project is supported by the Institute for Bioinformatics Research and Development of the Japan Science and Technology Agency, the 21st Century COE program ‘Genome Science’, and a grant-in-aid for scientific research on the priority area ‘Comprehensive Genomics’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The computational resource was provided by the Bioinformatics Center, Institute for Chemical Research, Kyoto University. Funding to pay the Open Access publication charges for this article was provided by the grant-in-aid for scientific research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moriya Y, Itoh M, Okuda S, Yoshizawa A, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashimoto K, Goto S, Kawano S, Aoki-Kinoshita KF, Ueda N, Hamajima M, Kawasaki T, Kanehisa M. KEGG as a glycome informatics resource. Glycobiology. 2006;16:63R–70R. doi: 10.1093/glycob/cwj010. [DOI] [PubMed] [Google Scholar]
- 5.Kawano S, Hashimoto K, Miyama T, Goto S, Kanehisa M. Prediction of glycan structures from gene expression data based on glycosyltransferase reactions. Bioinformatics. 2005;21:3976–3982. doi: 10.1093/bioinformatics/bti666. [DOI] [PubMed] [Google Scholar]
- 6.Minowa Y, Araki M, Kanehisa M. Comprehensive analysis of distinctive polyketide and nonribosomal peptide structural motifs encoded in microbial genomes. J. Mol. Biol. 2007;368:1500–1517. doi: 10.1016/j.jmb.2007.02.099. [DOI] [PubMed] [Google Scholar]
- 7.Kotera M, Okuno Y, Hattori M, Goto S, Kanehisa M. Computational assignment of the EC numbers for genomic-scale analysis of enzymatic reactions. J. Am. Chem. Soc. 2004;126:16487–16498. doi: 10.1021/ja0466457. [DOI] [PubMed] [Google Scholar]
- 8.Oh M, Yamada T, Hattori M, Goto S, Kanehisa M. Systematic analysis of enzyme-catalyzed reaction patterns and prediction of microbial biodegradation pathways. J. Chem. Inf. Model. 2007;47:1702–1712. doi: 10.1021/ci700006f. [DOI] [PubMed] [Google Scholar]
- 9.Limviphuvadh V, Tanaka S, Goto S, Ueda K, Kanehisa M. The commonality of protein interaction networks determined in neurodegenerative disorders (NDDs) Bioinformatics. 2007;23:2129–2138. doi: 10.1093/bioinformatics/btm307. [DOI] [PubMed] [Google Scholar]