Abstract
ChemBank (http://chembank.broad.harvard.edu/) is a public, web-based informatics environment developed through a collaboration between the Chemical Biology Program and Platform at the Broad Institute of Harvard and MIT. This knowledge environment includes freely available data derived from small molecules and small-molecule screens and resources for studying these data. ChemBank is unique among small-molecule databases in its dedication to the storage of raw screening data, its rigorous definition of screening experiments in terms of statistical hypothesis testing, and its metadata-based organization of screening experiments into projects involving collections of related assays. ChemBank stores an increasingly varied set of measurements derived from cells and other biological assay systems treated with small molecules. Analysis tools are available and are continuously being developed that allow the relationships between small molecules, cell measurements, and cell states to be studied. Currently, ChemBank stores information on hundreds of thousands of small molecules and hundreds of biomedically relevant assays that have been performed at the Broad Institute by collaborators from the worldwide research community. The goal of ChemBank is to provide life scientists unfettered access to biomedically relevant data and tools heretofore available primarily in the private sector.
INTRODUCTION
ChemBank v1.0 was initiated as a National Cancer Institute (NCI)-sponsored activity within the Initiative for Chemical Genetics (ICG), originally at Harvard's Institute of Chemistry and Cell Biology. The evolving interest of the NCI in sponsoring chemical-genetic research has been reported (1), as has the evolution of ICG as a public research effort dedicated to accelerating the discovery of cancer-relevant small-molecule probes (2). At present, ChemBank v2.0 (hereafter referred to as ChemBank) represents an evolving collaboration between the Chemical Biology Program and Chemical Biology Platform at the Broad Institute of Harvard and MIT, including interactions with academic synthetic chemists and biologists interested in high-throughput, small-molecule screening approaches.
ChemBank houses chemical structures and names, calculated molecular descriptors, human-curated biological information regarding small molecule activities, raw experimental results from high-throughput biological assays, and extensive metadata describing screening experiments. While there are many other publicly available small-molecule and drug databases [ChEBI (3), DrugBank (4), PubChem (5) and ZINC (6), among others], ChemBank is unique in three important ways: (i) its dedication to the storage of raw screening data; (ii) its rigorous definition of screening experiments in terms of statistical hypothesis testing; and (iii) its hierarchical metadata-based organization of related assays into screening projects. Moreover, the ChemBank website is more than a simple data repository; it contains tools for visualization and analysis of small-molecule results, including raw and normalized high-throughput screening (HTS) data and chemical-genetic profiles. Data sets may be manipulated within ChemBank or downloaded for use with external analysis tools. ChemBank is a powerful knowledge repository and analysis environment for chemists and biologists alike.
RESULTS
ChemBank infrastructure
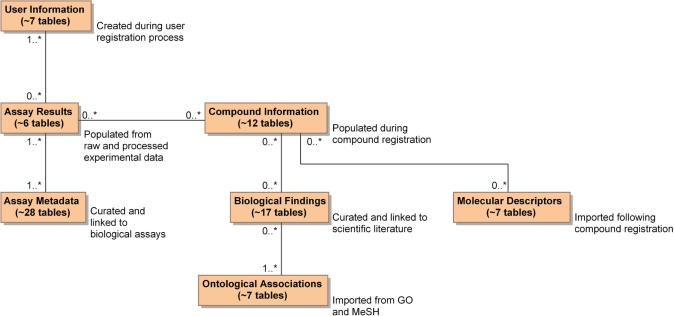
The ChemBank database consists of 95 tables segmented into seven logical groups representing compound information, molecular descriptors, assay results, assay metadata, biological findings, ontological associations and user information (Figure 1). Data from over 2500 high-throughput biological assays from 188 screening projects currently reside in ChemBank, with new assays loaded quarterly. ChemBank houses information on 1.7 million compound samples, representing more than 1.2 million unique small-molecule structures, with over 300 calculated molecular descriptor values for each molecule. Over 1000 proteins, 500 cell lines and 70 species are associated with the assays. ChemBank data are stored and searched using an Oracle 10g (Oracle Corporation; Redwood Shores, CA, USA) relational database extended with the DayCart Oracle cartridge (Daylight Chemical Information Systems; Aliso Viejo, CA, USA), which is used for molecule substructure and similarity searches.
Figure 1.
Conceptual summary of ChemBank schema. Logical illustration of ChemBank data model, in which 95 tables are organized into groups representing components of the chemical biology research enterprise. Each box represents several actual database tables, as indicated, and pseudocardinality relationships between boxes are meant to convey conceptual relationships, rather than the more complex cardinality relationships that relate the actual tables.
Data access and optional registration
Anyone with Internet access can use ChemBank, without registering with a username and password, using the ‘Enter as a Guest’ button. However, registration is highly encouraged of all users, as guest users are not permitted to export data from ChemBank. Registered users of ChemBank may download data directly from their search results using the ‘[export …]’ hyperlinks on several ChemBank webpages. Downloadable result files are exported in tab-delimited text format for maximum flexibility of use in other applications. Simple Object Access Protocol (SOAP) web-service access is provided for much of the data in ChemBank. Service calls exist to list all projects and assays within ChemBank, as well as assay plates and well measurements. There are also molecule services allowing similarity and substructure searches of molecules within ChemBank. The Web-service Definition Language (WSDL) files for these services can be found on the ChemBank website (http://chembank.broad.harvard.edu/webServices.htm).
As a repository of primary data from HTS experiments, ChemBank employs a data-embargo strategy to protect newly generated data for a specified period, and accordingly, ChemBank comprises two separate websites containing overlapping datasets. Public ChemBank (http://chembank.broad.harvard.edu/) contains small-molecule assay data that are more than one year old as well as molecular descriptors and published bioactivity annotation for registered small molecules. Data-Sharing-Agreement (DSA)-ChemBank contains all of the data content of Public ChemBank as well as small-molecule assay data that are less than one year old. Use of the latter database is restricted to scientists who have deposited compounds or performed screening experiments at the Broad Institute of Harvard and MIT and who have signed a DSA (http://www.broad.harvard.edu/chembio/sci/screen/facil/DataSharingAgreement.pdf). The DSA stipulates conditions of participation, such as shared authorship, intellectual property and expectations for sharing follow-up data. All scientists who have signed the agreement may browse the entire contents of DSA-ChemBank and renew their agreements annually for continued access to this resource. Registration for this version of ChemBank is required, and usernames and passwords are assigned by the ChemBank team upon receipt of the signed agreement. DSA-ChemBank users log in using their assigned username and password at a different URL (http://chembank-dsa.broad.harvard.edu/).
ChemBank queries, molecular properties and assay organization
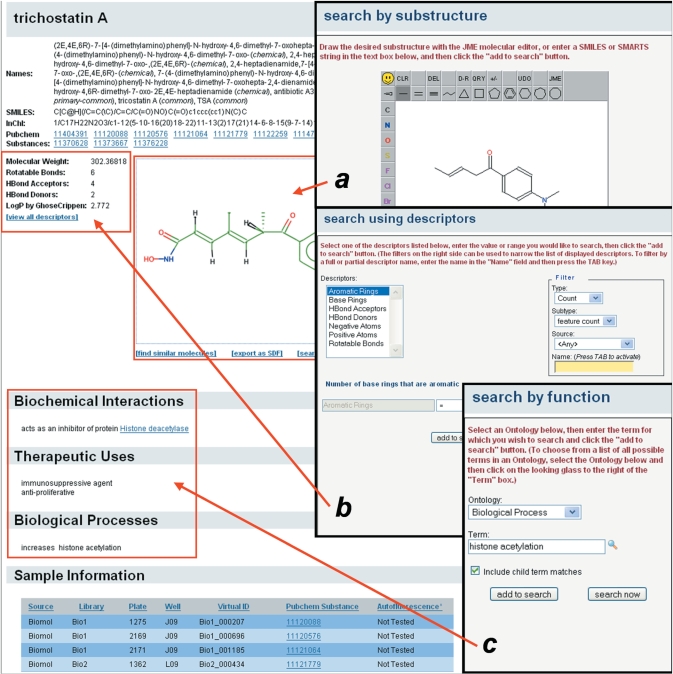
ChemBank has multiple search interfaces available to the user to enable both simple and complex queries. Users can search for small molecules using properties such as presence of a substructure (Figure 2a), calculated molecular descriptors (Figure 2b) or association with a curated biological activity (Figure 2c). Results from such searches lead the user to a ‘Molecule Display’ view, which shows basic information about the small molecule, including names, structure, chemical descriptors, biological activity and screening test instances (Figure 2, background).
Figure 2.
ChemBank offers multiple routes to find chemical information. Search tools allowing structure drawing (28) for substructure or similarity searches (a), selection of calculated molecular descriptors (b) and selection of term-based bioactivity annotations (c), each provide avenues to find individual molecules or sets of molecules in ChemBank. The ChemBank ‘Molecule Display’ webpage (background) provides detailed information about each molecule, including structure, names, molecular descriptors, biological annotations, sample information and screening instances.
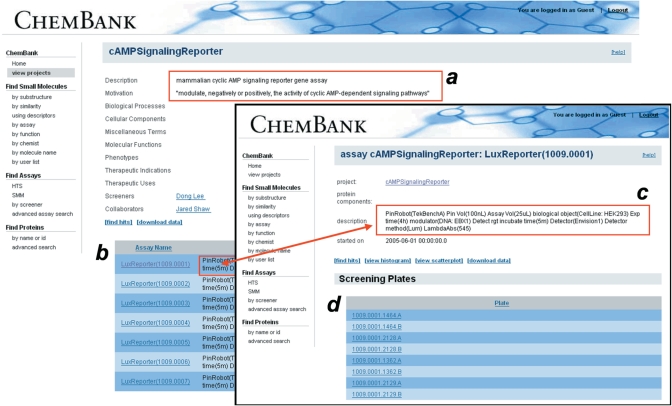
Users may also search for specific screening assays by searching by assay or project names, by individual screeners or their home institution, by assay type or by the species (e.g. of a cell line) under investigation in a particular assay. Text searches within screening project descriptions are also supported. Assay search result webpages link to both assay and screening project information; a project is a grouping of assays under a single biological motivation. Details of an assay or screening project are displayed on their respective webpages, including user information and assay metadata (Figure 3).
Figure 3.
Relationship of ChemBank ‘View Project’ and ‘View Assay’ webpages. Screenshots of representative screening project and assay (inset) webpages. Emphasis (red boxes, arrow) has been added to highlight key information, including project description and motivation (a), individual assays within project (b), detailed description (shared by both webpages) of assay protocol (c) and individual screening plates within assay (d).
Simple queries can be combined to generate complex, multi-criterion searches. ChemBank has an extensive multi-criterion search interface, which allows searches to be constructed interactively. This interface supports modification of prior search criteria or addition of new criteria, and can be used even after initial search results are returned. Additionally, data analysis on a single or combination of screens housed within ChemBank can be performed and visualized as detailed in the following section. More information on conducting ChemBank searches and webpages can be found in the Help section (http://chembank.broad.harvard.edu/details.htm?tag=Help).
ChemBank standard analysis model and visualizations
ChemBank houses both raw and normalized experimental results from many HTS and small-molecule microarray (SMM) (7) projects. These datasets are the foundation for a data-analysis model specifically equipped to accumulate rich profiles of small-molecule performance across multiple, diverse biological assays. One of the primary requirements of this approach is a data-analysis strategy for small-molecule performance that affords normalized values independent of both the screening technology platform and the specific biological question under investigation. ChemBank users are not restricted to this standard analysis paradigm; rather, each of the ChemBank data-visualization tools affords access to multiple data types along the analysis process, including raw data, background-subtracted data and replicate-handled data.
ChemBank seeks to provide cross-sectional analysis and multi-assay performance profiles (8–14). Therefore, we supplemented ChemBank raw screening data with normalized data using an error model with several salient features. First, it renders the results of multiple parallel assays formally comparable, regardless of the original signal amplitudes or units of measurement. Second, it introduces no assumptions about the strength of signal required to call screening positives. This condition rules out the use of arbitrary thresholds such as 2-fold induction or 50% inhibition (15,16). Third, it does not rely on global assumptions about the compound collection, particularly the assumption that most compounds will be inert in any given assay (17,18).
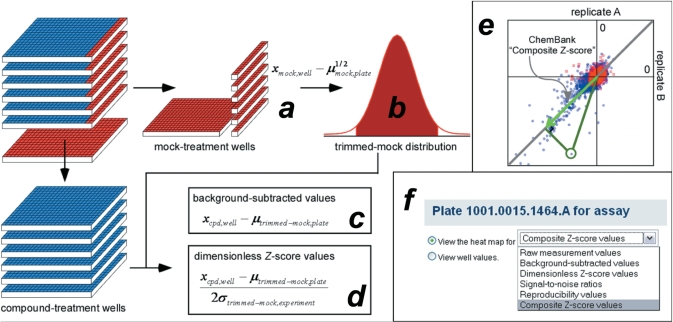
These three conditions suggested the use of only mock-treatment wells as a basis for well-to-well, plate-to-plate and experiment-to-experiment normalization. We reasoned that in any miniaturized assay of small-molecule performance, we can always define a mock-treatment condition that mirrors compound treatment in every way except for the presence of the compound. For example, in a typical HTS experiment, this condition is represented by the treatment of otherwise identical assay-well contents with the delivery vehicle, usually dimethylsulfoxide. We conceptualize our scoring method in terms of a formal hypothesis test comparing a compound-treatment with its associated mock-treatment distribution, and thus seek the most reliable estimates of the statistical parameters associated with this test. In an ideal world, one would simply make many measurements of a compound's performance in a particular assay system, and many measurements of the corresponding vehicle mock-treatment condition in that assay system. In such a case, our primary score for a compound would be similar to the Z' statistic for evaluating HTS methods during assay development (15,19). Since making a statistically meaningful number of measurements of each compound's performance is not practical, we estimate the parameters of the compound-treatment distribution from the parameters of the mock-treatment distribution. At present, ChemBank uses a constant-error assumption for plate-reader assays, and a variable error assumption for small-molecule microarrays (see subsequent paragraph).
To account for plate-to-plate variation in signal (15,16), the median raw value of mock-treatment signals on a given assay plate is subtracted from each mock-treatment value on the same plate (Figure 4a), providing a zero-centered distribution of mock-treatment measurements for each plate in one experiment. In ChemBank, a screening experiment is defined as a collection of distinct probe source plates (e.g. a multi-microtiter plate library) exposed to the same assay conditions at the same time. Next, the population of zero-centered measurements for all mock-treatment wells in the experiment are collected together, and values failing Chauvenet's criterion (20) for this overall distribution are discarded (Figure 4b), to protect against edge effects and other systematic artifacts known to present a technical problem for microtiter plate experiments (21,22). The remaining mock-treatment measurements are used to normalize each compound-treatment well measurement independently, first subtracting the mean of remaining mock-treatment wells on the same plate to obtain background-subtracted values (Figure 4c), then dividing by the twice the standard deviation of mock-treatment wells in the same experiment to obtain dimensionless Z-scores (Figure 4d). (Note that using twice the standard deviation is a consequence of a constant-error assumption for microplate assays described in the preceding paragraph; a more general solution allows the error estimate for positive signals to depend on signal strength, e.g., the scale factor used for small-molecule microarray experiments is very close to [1 + Isignal/Imock], as determined empirically using control arrays.) Thus, each compound well receives an algebraically signed Z-score corresponding to the number of standard deviations it fell above or below the mean of a well-defined mock-treatment distribution. Finally, as most screening experiments deposited in ChemBank were performed in two technical replicates (A and B), these replicates were combined to produce a Composite Z-score (Figure 4e) by scaling the vector [ZA,ZB] by the cosine correlation with a vector corresponding to ‘perfect reproducibility’ (i.e. equal Z-scores in both replicates). This overall normalization procedure is similar to that described in earlier work at the Broad Institute (9,12,23), and forms the basis for many of the ChemBank data visualizations available to users of the database (Figure 4f).
Figure 4.
ChemBank standard data-analysis model for high-throughput small-molecule screens. All raw small-molecule assay results in ChemBank are further processed by comparing each measurement with the collection of mock-treatment well measurements performed in the same screening experiment. Median values from mock-treatment wells on the same plate are used in an initial zero-centering step (a), after which the distribution of mock-treatment measurements for the entire experiment is trimmed to eliminate systematic artifacts (b). Trimmed mock-treatment measurements are used to normalize assay performance by first subtracting the mean of trimmed mock-treatment measurements on the same plate to give ‘background-subtracted values’ (c), then dividing by twice the standard deviation of trimmed mock-treatment measurements for the entire experiment to give ‘dimensionless Z-score values’ (d). Replicate handling is performed by cosine correlation of the replicate pair (for screens done in duplicate) of ‘dimensionless Z-score values’ for each compound with a simple prior model of ‘perfect reproducibility’, to yield a ‘Composite Z-score value’ (e) that represents the final primary screening result. The ChemBank web interface provides access to raw and processed data types appropriate for each of its visualization tools (f).
Currently, there are four available screening data visualizations in ChemBank. A simple heatmap visualization for assay plates corresponds to the microplate layout in a screening experiment (Figure 5a). For statistical analysis of results from multiple assay plates comprising a single HTS experiment, ChemBank offers both histogram (Figure 5b) and scatterplot (Figure 4e) views. Finally, multi-assay visualization is possible via the ‘Feature Visualization’ webpage (Figure 5c) which is generated by implementation of the standard analysis model to provide a heatmap representing ChemBank ‘Composite Z-score’ values for a series of assays and compounds. These visualization tools can be used in conjunction with the structure similarity, substructure matching, and molecular descriptor filtering capabilities described previously to perform structure–activity relationship analyses (Figure 5d). Taken together, these search and visualization tools represent an implementation of chemical-genetic profiling and cheminformatic analysis capabilities within ChemBank.
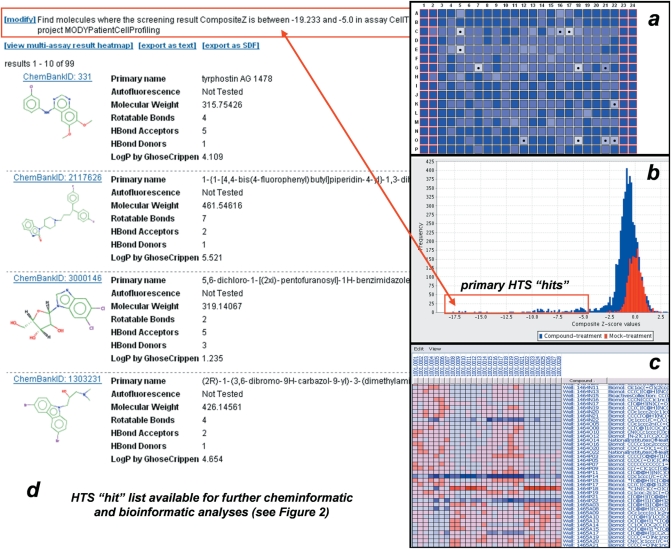
Figure 5.
Illustration of ChemBank visualizations and linking activities with chemical information. Screening data, including raw measurements, in ChemBank are addressable by exact plate and well position in assay plates (a), and statistical data representing outcomes (b) can be reviewed at the level of raw or normalized data. A multi-assay analysis capability takes advantage of the standard analysis procedure (Figure 4) to display the performance of such similar compounds in multiple assays to which each has been exposed (c). Each of these capabilities can be combined with structure and annotation-based search capabilities to provide cheminformatic analysis of molecules scoring as ‘hits’ in biological assays (d).
Data curation and annotation
Data curation activities for ChemBank fall into two general categories: 1) annotation of information regarding small molecules, and 2) annotation of information regarding small-molecule assays. Annotations regarding small-molecule effects on biological systems are generated from the primary literature. Hyperlinks to other databases such as Entrez Gene (24), GO (25), MeSH (http://www.nlm.nih.gov/mesh/meshhome.html) and PubMed (5), are collected manually during curation activities.
Small molecules and clinically used drugs often have multiple names; some drugs have dozens of different brand and generic names. ChemBank names are curated from multiple sources including public databases, the United States Pharmacopeia (USP) Dictionary, the World Health Organization (WHO) International Non-proprietary Name database (http://mednet.who.int/public/default.aspx?c=1f216b1a-c080-46a1-9a39-c33717387926), and chemical vendor websites to provide an exhaustive list of names and synonyms for molecules.
For a subset of the bioactive small molecules in ChemBank, activity annotations from the biological literature have been added. Bioactivity annotations (see also Figure 2) are divided into four categories: (i) Biochemical Interactions, which indicate protein molecular targets or GO molecular functions (25) affected by the small molecule; (ii) Therapeutic Indications, which specify what diseases (i.e. MeSH terms) a small molecule is used to treat or manage; (iii) Therapeutic Uses, terms from an internally derived vocabulary that indicates what type of clinical or biological activity a small molecule possesses; and (iv) Biological Processes, which indicate GO biological processes affected by the small molecule. PubMed IDs (5) corresponding to these citations have been captured, but are not yet exposed to the user and GO terms (Biochemical Interactions and Biological Processes) describing the effect(s) of the small molecule (e.g. ‘increases apoptosis’) are displayed but do not currently connect directly to the GO website (http://amigo.geneontology.org).
Annotation surrounding biological assays is collected as field-based metadata using internal controlled vocabularies, and is used to describe screening projects, experiments, and assay plates. Metadata associated with a screening project express the biological motivation and are displayed on the ‘View Project’ webpage; metadata associated with the details of a particular assay (instantiated as a screening ‘experiment’; see above) are displayed on the ‘View Assay’ webpage (see also Figure 3).
For many of the most frequently screened small molecules in ChemBank, annotation of autofluorescence activity is available to reduce false positives by providing automated filtering in addition to detailed information about the optical properties of the compounds. Annotation includes a binary filter of autofluorescence, a plot of the range of wavelengths above a signal threshold, a plot of the shape of the spectra and a contour plot showing the autofluorescence over a range that includes wavelengths of common filter pairs. Additional details about autofluorescence detection and data-analysis methods are available on the ChemBank website (http://chembank.broad.harvard.edu/details.htm?tag=Help#autoFluorMethodology).
DISCUSSION
ChemBank is a unique knowledge and analysis environment for small molecules and high-throughput small-molecule assays. It provides calculated molecular descriptors, experimental assay measurements, and literature-based annotation for small molecules, allowing integration of both new and established information in a single, public resource. Other public small-molecule databases exist, and the following examples are meant to be illustrative, not comprehensive. BindingDB (26) curates small-molecule binding affinities for protein targets from the literature. KEGG LIGAND (27) and its associated entry points seek to catalog and integrate chemical structures, biochemical reactions, and biological information into a single database. ZINC (6) is a database of three-dimensional compound structures intended for virtual screening applications. These products and the many other examples available serve to illustrate that each project has a different focus, and each is designed accordingly.
PubChem (5) is a public database, created by the NIH, that is most similar in aims and data content to ChemBank, but several important differences exist between ChemBank and PubChem (Table 1). Both databases house small-molecule structures and screening data, but ChemBank data are generated and annotated internally. PubChem is a deposition database, and relies on submission of data and structures from outside sources. Importantly, ChemBank houses raw screening data, including assay plate position information, and applies a standard data-analysis procedure to all datasets. PubChem does not require raw data or plate position information from submitters, and as such, interpretations of outcomes may be limited to those interpretations supplied by the submitting organization. ChemBank uses controlled vocabularies to capture metadata, and especially provides for hierarchical organization of assays into projects; PubChem captures data and protocols in free text, and organizes data by submission, rather than grouping assays by common biological motivation. The primary strengths of PubChem are its very useful links to other Entrez databases and its capability to leverage the powerful Entrez search engine. However, although both databases house general small-molecule information and bioassay data, we believe that storage of plate locations and raw screening data, field-based metadata, and the standard experiment definition in ChemBank afford some distinct advantages over PubChem. In particular, we believe that ChemBank will be especially effective in revealing unrecognized small-molecule performance relationships because of its support of statistically rigorous cross-sectional analyses that harness the collective power of all experiments in the database (i.e. analyses of data derived from many different types of screens and small molecules).
Table 1.
Content and feature comparison between ChemBank v2.0 and PubChem
| Database content/feature | ChemBank v2.0 | PubChem |
|---|---|---|
| Chemical structure | >1.2 million unique | >10 million unique |
| Molecule names/synonyms | IUPAC and manually curated names/synonyms | IUPAC and depositor-supplied synonyms |
| Molecular descriptors | 36 searchable plus >300 displayed calculated properties and descriptors per molecule | 18 calculated chemical properties, atom types and stereochemistry flags per molecule |
| Literature annotations | Manually curated; connected to controlled vocabularies | Extensive linking to Entrez databases |
| Metadata describing screens | Extensive field-based metadata collected (some displayed); standardized terms connected to controlled vocabularies | Depositor-supplied comments; primarily free-text, no standard controlled vocabularies for assay components |
| Biological assay data | Raw data required with plate locations (except legacy ChemBank v1. × data sets); standard data-analysis model | ∼15% of assays display raw data; no standardized analysis performed beyond submitter interpretations |
| Screening assay analysis and visualization tools | Plate-map, histogram, scatterplot and multi-assay heat-map visualizations | Structure-activity relationship analyses; clustering visualizations |
| Public data submission | Required for Broad Institute screening center; limited external submissions | Required for Molecular Libraries Screening Centers; others may submit voluntarily |
By committing itself to storing raw screening data, defining screening experiments in a rigorous manner, and organizing screening experiments hierarchically using metadata, ChemBank provides a unique opportunity to the scientific community, even beyond computational scientists. ChemBank provides access to large volumes of data annotating chemical entities with measured outcomes, and the commitment to storage of raw data ensures that these measurements are available for data-mining activities. In this sense, ChemBank is an important social experiment in science, in that it permits an analysis of whether the interpretation of screening data by the investigators performing small-molecule screens indeed reveal all the value in their performance. Alternatively, cross-sectional analysis of results deposited by multiple independent investigators may reveal outcomes not apparent to participants in any individual screening project. To that end, ChemBank allows customization of options for viewing chemical-genetic profiles for the resulting molecules using any associated assay data, and as such, allows users to exploit this assembled information to support discovery and experimentation in chemical biology research. The open, standardized data-sharing environment of ChemBank is geared toward rapid discovery of novel therapeutic candidates and a deep understanding of biological systems.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the support of the National Cancer Institute's Initiative for Chemical Genetics (N01-CO-12400), the NIH Road Map's Exploratory Center for Cheminformatics Research (P20-HG003895), and the NIGMS Center for Chemical Methodologies and Library Development (P50-GM069721, including supplemental funds). We further acknowledge the Broad Institute's IT/Systems group for infrastructural support, and numerous contract workers who have helped to augment ChemBank's contents: Natalia Balabi, Jose A. Fernandez, Cynthia A. Saraceni-Richards, Karen Rose, Laura Selfors, Nurgees Sulthan-Banu, Brian Weiner and Angela Zuniga-Meyer. Finally, we are indebted to the following investigators for early modeling efforts, helpful discussions and community participation during ChemBank development: Elton Dean, David DeCaprio, Jay Duffner, Scott Eliasof, Joshua Forman, Annaliese Franz, Julie Gorenstein, Stephen Haggarty, Eugenia Harris, Angela Koehler, Andrew Lach, Justin Lamb, Julia Lamenzo, Ralph Mazitschek, John McGrath, Olivia McPherson, Jared Shaw, Stanley Shaw, Lynn Verplank and Bridget Wagner. Funding to pay the Open Access publication charges for this article was provided by Initiative for Chemical Genetics (NO1-CO-12400).
Conflict of interest statement. We report that M.S. provided consultancy services, the subject(s) of which were unrelated to the ChemBank project, to Daylight Chemical Information Systems while participating in ChemBank development.
REFERENCES
- 1.Strausberg RL, Schreiber SL. From knowing to controlling: a path from genomics to drugs using small molecule probes. Science. 2003;300:294–295. doi: 10.1126/science.1083395. [DOI] [PubMed] [Google Scholar]
- 2.Tolliday N, Clemons PA, Ferraiolo P, Koehler AN, Lewis TA, Li X, Schreiber SL, Gerhard DS, Eliasof S. Small molecules, big players: the National Cancer Institute's Initiative for Chemical Genetics. Cancer Res. 2006;66:8935–8942. doi: 10.1158/0008-5472.CAN-06-2552. [DOI] [PubMed] [Google Scholar]
- 3.Brooksbank C, Cameron G, Thornton J. The European Bioinformatics Institute's data resources: towards systems biology. Nucleic Acids Res. 2005;33:D46–D53. doi: 10.1093/nar/gki026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irwin JJ, Shoichet BK. ZINC - a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duffner JL, Clemons PA, Koehler AN. A pipeline for ligand discovery using small-molecule microarrays. Curr. Opin. Chem. Biol. 2007;11:74–82. doi: 10.1016/j.cbpa.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 8.Fliri AF, Loging WT, Thadeio PF, Volkmann RA. Biological spectra analysis: linking biological activity profiles to molecular structure. Proc. Natl Acad. Sci. USA. 2005;102:261–266. doi: 10.1073/pnas.0407790101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franz AK, Dreyfuss PD, Schreiber SL. Synthesis and cellular profiling of diverse organosilicon small molecules. J. Am. Chem. Soc. 2007;129:1020–1021. doi: 10.1021/ja067552n. [DOI] [PubMed] [Google Scholar]
- 10.Haggarty SJ, Clemons PA, Schreiber SL. Chemical genomic profiling of biological networks using graph theory and combinations of small molecule perturbations. J. Am. Chem. Soc. 2003;125:10543–10545. doi: 10.1021/ja035413p. [DOI] [PubMed] [Google Scholar]
- 11.Kauvar LM, Higgins DL, Villar HO, Sportsman JR, Engqvist-Goldstein A, Bukar R, Bauer KE, Dilley H, Rocke DM. Predicting ligand binding to proteins by affinity fingerprinting. Chem. Biol. 1995;2:107–118. doi: 10.1016/1074-5521(95)90283-x. [DOI] [PubMed] [Google Scholar]
- 12.Kim YK, Arai MA, Arai T, Lamenzo JO, Dean EF, III, Patterson N, Clemons PA, Schreiber SL. Relationship of stereochemical and skeletal diversity of small molecules to cellular measurement space. J. Am. Chem. Soc. 2004;126:14740–14745. doi: 10.1021/ja048170p. [DOI] [PubMed] [Google Scholar]
- 13.Melnick JS, Janes J, Kim S, Chang JY, Sipes DG, Gunderson D, Jarnes L, Matzen JT, Garcia ME, et al. An efficient rapid system for profiling the cellular activities of molecular libraries. Proc. Natl Acad. Sci. USA. 2006;103:3153–3158. doi: 10.1073/pnas.0511292103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaharevitz DW, Holbeck SL, Bowerman C, Svetlik PA. COMPARE: a web accessible tool for investigating mechanisms of cell growth inhibition. J. Mol. Graph Model. 2002;20:297–303. doi: 10.1016/s1093-3263(01)00126-7. [DOI] [PubMed] [Google Scholar]
- 15.Brideau C, Gunter B, Pikounis B, Liaw A. Improved statistical methods for hit selection in high-throughput screening. J. Biomol. Screen. 2003;8:634–647. doi: 10.1177/1087057103258285. [DOI] [PubMed] [Google Scholar]
- 16.Gunter B, Brideau C, Pikounis B, Liaw A. Statistical and graphical methods for quality control determination of high-throughput screening data. J. Biomol. Screen. 2003;8:624–633. doi: 10.1177/1087057103258284. [DOI] [PubMed] [Google Scholar]
- 17.Zhang JH, Chung TD, Oldenburg KR. Confirmation of primary active substances from high throughput screening of chemical and biological populations: a statistical approach and practical considerations. J. Comb. Chem. 2000;2:258–265. doi: 10.1021/cc9900706. [DOI] [PubMed] [Google Scholar]
- 18.Zhang JH, Wu X, Sills MA. Probing the primary screening efficiency by multiple replicate testing: a quantitative analysis of hit confirmation and false screening results of a biochemical assay. J. Biomol. Screen. 2005;10:695–704. doi: 10.1177/1087057105279149. [DOI] [PubMed] [Google Scholar]
- 19.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 20.Bevington PR, Robinson DK. Data reduction and error analysis for the physical sciences, 2nd. Boston, MA: McGraw-Hill; 1991. [Google Scholar]
- 21.Kelley BP, Lunn MR, Root DE, Flaherty SP, Martino AM, Stockwell BR. A flexible data analysis tool for chemical genetic screens. Chem. Biol. 2004;11:1495–1503. doi: 10.1016/j.chembiol.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 22.Root DE, Kelley BP, Stockwell BR. Detecting spatial patterns in biological array experiments. J. Biomol. Screen. 2003;8:393–398. doi: 10.1177/1087057103254282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly KA, Clemons PA, Yu AM, Weissleder R. High-throughput identification of phage-derived imaging agents. Mol. Imaging. 2006;5:24–30. [PubMed] [Google Scholar]
- 24.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gene Ontology C. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ertl P, Jacob O. WWW-based chemical information system. J. Mol. Struct. Theochem. 1997;419:113. [Google Scholar]