Abstract
Affymetrix exon arrays aim to target every known and predicted exon in the human, mouse or rat genomes, and have reporters that extend beyond protein coding regions to other areas of the transcribed genome. This combination of increased coverage and precision is important because a substantial proportion of protein coding genes are predicted to be alternatively spliced, and because many non-coding genes are known also to be of biological significance. In order to fully exploit these arrays, it is necessary to associate each reporter on the array with the features of the genome it is targeting, and to relate these to gene and genome structure. X:Map is a genome annotation database that provides this information. Data can be browsed using a novel Google-maps based interface, and analysed and further visualized through an associated BioConductor package. The database can be found at http://xmap.picr.man.ac.uk.
INTRODUCTION
Approximately 50% of human genes are predicted to be alternately spliced (1,2), and up to 90% of the entire genome to be transcribed (3,4). Recent work has shown that many non-coding genes are of direct functional significance, that there is strong conservation in regions once characterized as simply ‘junk’ DNA, and that there are many putative novel exons and transcripts in relatively well-characterized protein coding regions (4,5). Many loci also contain multiple interwoven and overlapping genes, possibly translated in different reading frames, adding additional layers of complexity (5). Together, these issues raise a number of challenges for gene expression studies. In particular, it is desirable both to use multiple reporters for individual genes in order to pursue splicing events and to increase coverage to explore the less well-characterized regions of the genome. As knowledge of the complexities of gene expression increases, so does the demand for increasingly feature-dense arrays.
Advances in microarray technology are beginning to address these issues by offering significantly increased feature densities and a concomitant rise in the number of available reporters. The Affymetrix Human Exon 1.0ST array, for example uses ∼6.5 million probes, comprising ∼1.4 million probesets, to target ∼1.2 million individual exons, and similar arrays exist for mouse and rat. The aim is to target every known and predicted exon in the genome (6–8). These arrays offer a number of opportunities, but place significant challenges on the bioinformatics tools necessary to exploit them. In particular, sufficient annotation must be provided, for example to identify which exons, transcripts and genes are targeted by each probeset, and the levels of evidence used to define each of these features within the genome (7).
X:Map is a database designed to provide this information. It offers detailed annotation of the intron–exon structure of each gene, their mappings to known transcripts, and their location relative to Affymetrix exon array target sequences. Data can be explored using a fully interactive real-time scrollable browser, built using the Google Maps API (Figure 1). Other groups have also considered AJAX-based approaches to this problem, most notably GBrowse-AJAX (available on sourceforge.net). However, these are currently at the proof of concept/prototype stage.
Figure 1.
The X:Map genome browser. (a) Scrollable genome map. (b) Toolbar. (c) Hierarchical annotation of map features. (d) Context-dependent annotation.
An associated BioConductor (9) package, ‘exonmap’ (7), provides an interface between the database and R, a popular programming language for data analysis. X:Map is unique because although some of these data are provided by, for example, Affymetrix through their NetAffx (10) database, comprehensive mappings to the genome, filtering for match specificity and provision of the data in a form that supports both high-throughput and gene-centric analysis, are not available in an integrated form elsewhere.
DATA GENERATION AND DATABASE BUILDS
X:Map is built by searching, in silico, every 25-mer probe sequence represented on the microarray against the entire genome [downloaded from Ensembl (11)] and recording the location of each exact match in a relational database. The search strategy is discussed in ref. (12). Genome searches are performed because many of the probesets on the array target less well-characterized regions [e.g. GenScan (13) predictions], and because the large amount of ‘intergenic’ transcription observed means that it is necessary to consider each probe's potential to hybridize outside annotated genes. This is discussed further below. An additional search is also performed against a database of known transcript sequences (Ensembl cDNA data) to identify probes that cross exon boundaries; these would not be found at the genome level. This is all done via a series of Groovy (groovy.codehause.org) scripts initiated from a master Java process. Groovy was used because it allowed a much faster turnaround during prototyping and testing, while maintaining reasonable performance (Groovy scripts are compiled to Java before they are executed).
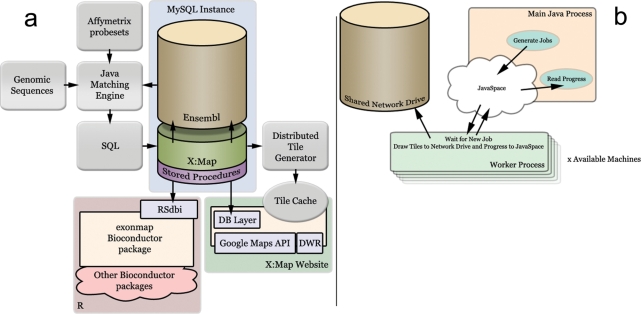
These data are used to populate a set of new tables in a local copy of Ensembl (11), indexed for speed of querying. An additional set of stored procedures are imported to provide a well-defined API for both the website and the BioConductor package. The Ensembl database does not support foreign keys, but these new tables link to the existing ones using the same unique identifiers as those used by Ensembl. The overall architecture of X:Map is shown in Figure 2a. In total, this phase of the build process takes ∼3 h for Ensembl v45 (human, mouse and rat combined) on a twin-Xeon 64 bit machine with 16 GB RAM running Red Hat Enterprise Linux.
Figure 2.
The X:Map build process. (a) Overview. The entire human genome is searched against the array's 25-mer probe sequences by the Java Matching engine. Results are stored in a relational database as a set of additional tables accompanying a local installation of Ensembl. The database is used both by an R client that supports analysis using BioConductor and a webserver that supports a Web 2.0/AJAX Genome Browser. The browser serves a set of pre-computed images generated offline by a coarse-grained parallel task (b). (b) Distributed computation of image tiles using JavaSpaces. Tile generation jobs are placed in a JavaSpace by a master process (Main Java Process). These are consumed by a series of worker processes distributed across a variety of Linux machines. All image data are written to a shared network drive.
The X:Map genome browser makes use of pre-drawn tiled images in order to improve run-time speed. These are generated in a second phase, managed as a coarse-grained parallel task distributed across a number of Linux servers and workstations (Figure 2b) using a producer/consumer model. In this way, spare cycles available within existing hardware are exploited. During this phase of the build, images are generated using a disparate set of 11 Linux servers and workstations in ∼3 days. Tiles are then processed to generate three alternate levels of magnification (∼5 days). In the end, for all three species, layers and zoom levels, this process generates 52 GB of image data, comprising 10.27 million individual image tiles.
THE DATA
The original Exon array annotation supplied by Affymetrix grouped probesets into three sets based on the reliability of the annotation used to define the putative genomic regions of interest. X:Map groups probesets based on Ensembl annotations, and currently provides data for Ensembl Genes, Ensembl ESTs (predictions based on EST data) and GenScan predictions (11). In this way, a direct mapping is formed between the probesets and the data sources used to provide the annotations found at the location the probeset matches the genome.
The arrays use a set of short 25-mer probes to target each feature of interest, together referred to as a ‘probeset’. The majority of exon array probesets contain four probes. Each vertical line in the background of Figure 1a represents a probe. It is coloured green if that sequence can match the genome in only one location, grey if it matches at more than one site. X:Map considers a probeset to be ‘multi-targeting’ if one or more of its probes matches at multiple genomic locations. X:Map also categorizes probesets according to whether they hit within an exon (‘exonic’), within a gene but outside an exon (‘intronic’) or between genes (‘intergenic’).
The relative proportions of probesets in each of these categories are summarized for human mouse and rat arrays in Table 1. It can be seen, therefore, that a substantial number of probesets on the array target outside known Ensembl genes, or within genes, but outside known exons, and that many of these match instead to EST or GenScan predictions. Data analysis is best performed with knowledge of these distinctions (7). Even though relatively few probesets (<10%) contain one or more probes capable of hybridizing to multiple sites, their identification is important because many are highly promiscuous and each may match multiple regions of interest (7).
Table 1.
Relative proportions of probesets in different categories for Human, Mouse and Exon arrays
| Human | Mouse | Rat | ||||
|---|---|---|---|---|---|---|
| Ensembl version | 45_36 g | 45_36 f | 45_34o | |||
| Total probeset hits | 1432150 | 1235873 | 1044729 | |||
| Multitarget | 129404 | 9% | 87181 | 7% | 71163 | 7% |
| Exonic | 301030 | 21% | 257421 | 21% | 222876 | 21% |
| Intronic probesets | ||||||
| all | 329007 | 23% | 284182 | 23% | 194941 | 19% |
| within EST exons | 18851 | 1% | 25303 | 2% | 5057 | <1% |
| within genscan | 16852 | 1% | 15683 | 1% | 15848 | 2% |
| Intergenic probesets | ||||||
| all | 650196 | 45% | 591475 | 48% | 537732 | 51% |
| within EST exons | 31913 | 2% | 37691 | 3% | 12653 | 1% |
| within genscan | 49236 | 3% | 48842 | 4% | 52031 | 5% |
Data were filtered to remove multi-target probesets and then matched against Ensembl genes. Intronic and intergenic probesets were then searched against Ensembl ESTs and Genscan predictions to assign matches to putative exons.
THE BROWSER
X:Map supports a novel genome browser (Figure 1). One of the issues of presenting genomic data is the need to represent structures such as genes that are many thousands of residues long, while being able also to represent other features, such as a probe hybridization location that are a few (25 bp) residues in length. This requires a browser that can not only offer a variety of different zoom levels but can also support swift navigation up- and downstream of the current location. Other important considerations are the need to maximize the amount of screen real estate available to present data, the potential for clutter if too many features are presented at the same time and a simple but capable search interface. X:Map, which is designed specifically for high-density microarrays was required to show for each gene, transcript and exon the probesets that match it, their specificity and for each probe, their locations of potential hybridization, and for each individual exon, its sequence. These requirements were addressed by using a visual map with multiple levels of zoom. In order to maximize the responsiveness of the browser this is implemented by generating a set of pre-drawn images (for the entire genome) at various levels of magnification. These are then stored on the server as a series of small, tiled, image files, which are managed by the client using the Google Maps API, which handles the task of requesting the appropriate tiles from the server, of assembling them into a larger contiguous image, as well as the scrolling and zooming of the map. Consequently, no computation is required to generate images at run-time. The result is a fully interactive map that can be scrolled dynamically, and with a user interface that, through the use of the Google Maps API, will be already familiar to many users. In order to maximize the amount of data that can be displayed, the genome is drawn horizontally (most monitors are wider than they are tall) and extends the full width of the browser window.
Data are presented as a set of transparent layers that can be added or removed from the image. By default, only the Ensembl Gene layer is shown; EST and GenScan predictions can be overlaid as required. Clearly, since the Ensembl annotation pipeline also makes use of ESTs and in silico predictions, there is a significant degree of overlap between these data, and statistical analyses using exonmap (see below) should take this non-independence into account. The tools menu (Figure 1b) can be hidden or shown by clicking on its tab. When a region of the map is selected, neighbouring features are listed in a selection box; selection from which causes information pertaining to that item to be displayed as a tree (Figure 1c). If the menu is hidden, the tab is animated (it briefly moves left and right a few pixels) providing a visual cue to draw attention to the fact that its state has changed. The hierarchy (Figure 1c) shows the nested relationships between genes, transcripts, exons, probesets, probes and their match locations. Selecting an item in the hierarchy causes contextual annotation to be displayed (Figure 1d) along with hyperlinks to external databases. Thus genes and transcripts are annotated with HUGO symbols, descriptions and hyperlinks to external databases such as Ensembl and GeneCards (14), exons with their reading frame and the appropriate fragment of genomic sequence, and probes and probesets with probe sequences and links to NetAffx (10). Icons are used to identify probesets that are annotated as non-specific, exonic, intronic and intergenic, both in the hierarchy and the annotation view. Detailed descriptions of the different icons and visual cues used within the browser can be found on the help pages accompanying the web site.
DATA ANALYSIS
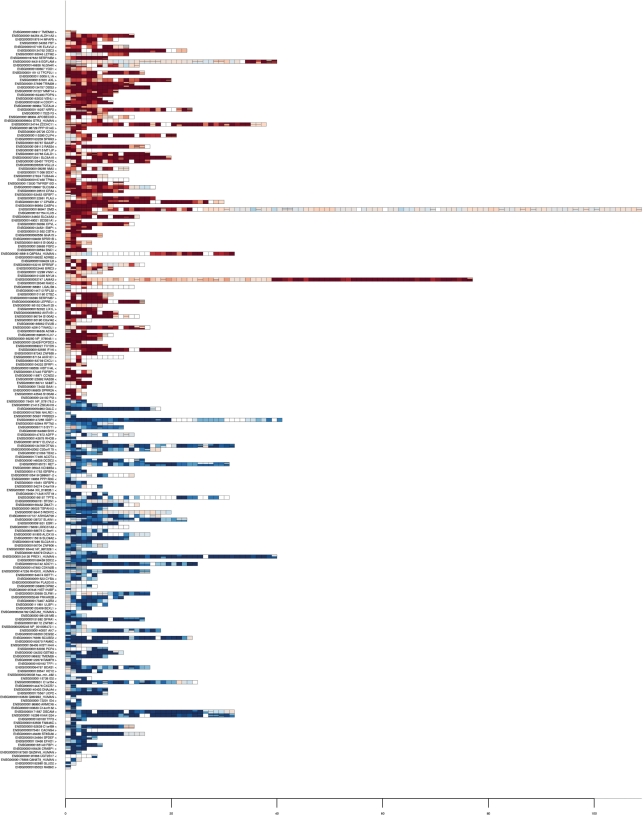
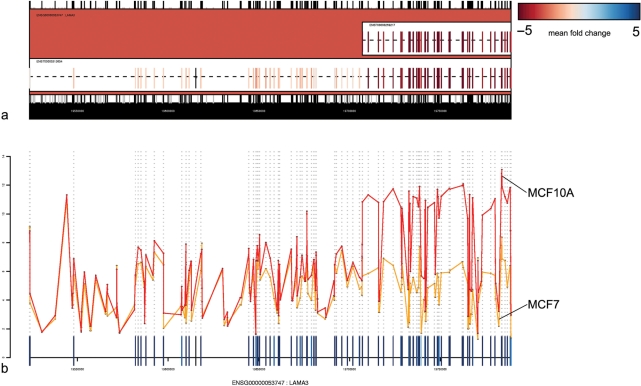
A BioConductor package, ‘exonmap’, can connect to the database (via the Rdbi package) in order to extract annotation data for use within a full statistical analysis environment. The package provides a series of functions allowing mappings to be made between probeset, exon, transcript and gene identifiers, and filterings to be performed to include/exclude exon, intron and intergenic probesets and those that are non-specific to the genome. Additional functions allow genome features to be retrieved according to physical location. Finally, a set of visualization functions can be used to map expression data onto genomic features (Figures 3 and 4).
Figure 3.
Genes selected as alternatively spliced between MCF7 and MCF10A cell lines, coloured by fold change. Each row corresponds to a gene, each rectangle, an exon. Exons are arranged in position order. Exons targeted by multiple probesets are drawn with these stacked horizontally within the exon. White rectangles correspond to exons with missing data. Fold change colouring ranges from 2−5 (intense blue; up in MCF7) to 25 (intense red; up in MCF10A).
Figure 4.
Microarray expression data mapped to the gene structure for LAMA3. (a) Two known isoforms are represented in Ensembl. Expression data are mapped onto these and coloured according to fold change between MCF7 and MCF10A. The pattern of expression across the length of the gene clearly corresponds to the known gene structure. (b) Line plot for the same data.
A possible workflow for data analysis using X:Map and exonmap is to first identify a set of differentially expressed probesets. This can be done using standard techniques developed for conventional microarrays. These are then mapped, using X:Map, to their target genes, and filtered for non-specificity due to multiple targeting. The result is a list of genes for which at least one probeset is differentially expressed.
Then, for each of these genes, it is possible to retrieve expression data for every targeting probeset. Fold change (e.g. splicing index), ANOVA (e.g. MIDAS) or variance-based approaches can then be used to characterize genes according to consistency in differential expression across their length (15,16). Figure 3, for example, was generated by using limma (17) to identify all probesets with statistically significant differential expression in a triplicate comparison between two cell lines, the human breast cancer cell line MCF7, and the non-tumorigenic breast epithelial cell line, MCF10A (N = 90 051; fold change threshold >2; adjusted P value = 0.05, Benjamini and Hochberg correction). This list was then filtered to remove multi-target probesets and those that did not map cleanly to exons (N = 49 326). Variance was calculated for each gene, and used to select the 200 most varying up- and downregulated genes. The dataset is described in detail in ref. (6). Gene expression can also be mapped to the transcript structure of individual genes. Figure 4a shows, for example, the gene LAMA3, with expression mapped to its transcript structure. A set of exons at the 3′ end of the gene appear differentially expressed with respect to the rest of the gene. This corresponds in location to a known short isoform. Figure 4b shows mean intensity for MCF7 and MCF10A plotted for the same gene against sequence position. It can be seen that the expression levels for both cell lines are similar at the 5′ end, but that expression levels increase in MCF10A at the 3′ end (rather than decreasing for MCF7). This provides strong evidence that the changes observed are due to over-expression of the known short isoform in MCF10A, rather than ‘under-expression’ of the 5′ end of the gene in MCF7.
FUTURE CHALLENGES
Currently, X:Map focuses on Affymetrix Exon arrays, however, many of the approaches can be generalized to other array types, and in particular, to tiling and SNP arrays. We intend to add these array types to X:Map, and also to extend the types of annotation represented within the database to include data types more pertinent for these arrays. An advantage of the approach taken by X:Map is that, since all visualization data are pre-computed, making more annotation available via the web-interface will not result in a loss of run-time performance. We are also exploring the possibility of adding an additional client-rendered layer to allow dynamic presentation of users’ own expression data alongside the existing map annotation.
AVAILABILITY
All database data can be downloaded from the X:Map website, exonmap from the BioConductor website and the MCF7 and MCF10A sample data from ref. (6).
ACKNOWLEDGEMENTS
This work was funded by Cancer Research UK. We are grateful to the BioConductor community for their considerable efforts and to David James for his help and encouragement with RMySQL. Exon array data were generated by the Cancer Research UK Affymetrix Service; we are also grateful to Siân Dibben for extensive testing and feedback using X:Map and exonmap. Funding to pay the Open Access publication charges for this article was provided by Cancer Research UK.
Conflict of interest statement. None declared.
REFERENCES
- 1.Modrek B, Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 2.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 4.ENCODE consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, et al. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 6.Okoniewski MJ, Hey Y, Pepper SD, Miller C. High correspondence between Affymetrix exon and standard expression arrays. Biotechniques. 2007;42:181–185. doi: 10.2144/000112315. [DOI] [PubMed] [Google Scholar]
- 7.Okoniewski MJ, Yates T, Dibben S, Miller CJ. An annotation infrastructure for the analysis and interpretation of Affymetrix exon array data. Genome Biol. 2007;8:R79. doi: 10.1186/gb-2007-8-5-r79. http://genomebiology.com/2007/8/5/R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardina P, Clark T, Shimada B, Staples M, Yang Q, Veitch J, Schweitzer A, Awad T, Sugnet C, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentleman RC, Carey VJ, Bates DJ, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, et al. Bioconductor Project Working Papers, Working Paper 1. 2004. Bioconductor: open software development for computational biology and bioinformatics. http://www.bepress.com/bioconductor/paper1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu G, Loraine AE, Shigeta R, Cline M, Cheng J, Valmeekam V, Sun S, Kulp D, Siani-Rose MA. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003;31:82–86. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leong HS, Yates T, Wilson C, Miller CJ. ADAPT: a database of Affymetrix probesets and transcripts. Bioinformatics. 2005;21:2552–2553. doi: 10.1093/bioinformatics/bti359. [DOI] [PubMed] [Google Scholar]
- 13.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics. 1998;14:656–664. doi: 10.1093/bioinformatics/14.8.656. [DOI] [PubMed] [Google Scholar]
- 15.Clark T, Schweitzer A, Chen T, Staples M, Lu G, Wang H, Williams A, Blume J. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Affymetrix. Affymetrix Whitepaper. 2005. Alternative transcript analysis methods for exon arrays. www.affymetrix.com. [Google Scholar]
- 17.Smyth G. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. New York: Springer; 2005. pp. 397–420. [Google Scholar]