Abstract
The hepatitis C virus (HCV) is a significant public health threat worldwide. The virus is highly variable and evolves rapidly, making it an elusive target for the immune system and for vaccine and drug design. Presently, ∼50 000 HCV sequences have been published. A central website that provides annotated sequences and analysis tools will be helpful to HCV scientists worldwide. The HCV sequence database collects and annotates sequence data, and provides them to the public via a website that contains a user-friendly search interface and a large number of sequence analysis tools, following the model of the highly regarded and widely used Los Alamos HIV database. The HCV website can be accessed via http://hcv.lanl.gov and http://hcv-db.org.
INTRODUCTION
The hepatitis C virus (HCV) has infected 4 million people in the United States and ∼170 million people worldwide. HCV infection is cleared in ∼25% of cases (1,2), and in the rest results in chronic infection. A recent study (3) estimated that lifetime HCV-associated mortality is around 1 in 8; a much larger number (an estimated 1 in 4) will develop cirrhosis of the liver. Most likely this number will be higher in less developed countries. With 170 million people infected worldwide, this means 20 million HCV-related deaths in the next few decades.
The infection has been known for decades and was previously called non-A non-B hepatitis, but the virus was not discovered until 1989 (4). It is spread through blood and blood products. Reliable tests have been available since soon after the discovery of the virus (5), but there are still occasional outbreaks in the Western world, either because the carrier is in the very early stage of the infection when antibodies are not yet detectable, or by trace amounts of virus, for example via kidney dialysis equipment (6). Worldwide, the epidemic is still spreading via contaminated blood and needles.
HCV is a positive-sense RNA virus with a genome of ∼9400 bases, which encodes a single polyprotein that is cleaved into four structural proteins (Core, Envelope 1 and 2 and p7) and six non-structural proteins named NS2-NS5B. It has been classified as a member of the genus hepacivirus. Hepaciviridae in turn are part of the family that includes Dengue, yellow fever and West Nile virus, with which it shares many structural features. However, the genetic distance between HCV and other flaviviruses is >50% over the entire genome (7). HCV is an extremely variable virus that forms polymorphic swarms of variants within the host. Worldwide, six different genotypes have now been defined, and each of those is subdivided into a total of more than 67 subtypes, of which 19 have been completely sequenced.
It is expected that both the generation of escape and resistance mutations and the high variability itself will complicate drug and vaccine design (8). In the closely related field of HIV research, there is a renewed interest in rational vaccine design, a relatively new discipline that attempts to define the optimal vaccine strain, possibly an artificially created one, that minimizes the differences from circulating strains while maximizing the immunogenicity of the reagent (9). Since for hepatitis C both drug and vaccine design are in their infancy, a database that allows researchers to study genetic variability by facilitating retrieval, alignment and analysis of all publicly available HCV sequences could play an important role in helping these efforts.
THE PURPOSE AND DESIGN OF THE DATABASE
The HCV database aims to be a resource for scientists working on HCV genetics, evolution, variability, and vaccine and drug design. The database is managed by biologists with extensive experience in sequence analysis, assisted by bioinformaticians and an editorial board consisting of international experts in the field of HCV research.
The backbone of the database is formed by the HCV sequences deposited in GenBank. New sequences are downloaded weekly, and the available ancillary information is extracted from the GenBank records. This information may include country, sampling year, isolate names, genotype and subtype, host species, etc. For most of the sequences (excluding those shorter than 150 nt), relevant annotation information is obtained from the associated publications and added to the database. As much as possible, sequences that do not have a genotype and subtype assigned are manually typed using phylogenetic analysis and BLAST searches. Annotation is being added continuously, both to newly downloaded sequences and to sequences already in the database.
Presently, annotation fields in the database include:
Sequence information
Genotype, subtype, start and stop coordinates relative to the reference strain HCV-H77, sampling country, -city, -date and -tissue.
Patient information
Health status, age, gender, ALT level, treatment and result, co-infection with HIV and hepatitis B, infection date, -country, -city, -route, and -outcome, HLA type, and epidemiological relations to other patients.
The information in the database can be accessed via a versatile but user-friendly search interface that allows searches on some 30 different fields, and lets the user automatically exclude sequences from non-human hosts, sequences from patent applications and sequences that have a close epidemiological relation (either from one patient or from a cluster of linked infections). The search results can be sorted and selected in various ways, and include an icon for each sequence that shows at a glance how long each sequence is and where in the genome it is located. A graphical overview showing which regions and which genotypes are included in the entire set of retrieved sequences can be generated (Figure 1). An important feature is the ability to search by genomic region, so the user can locate all sequences in the database that span (for example) E1 and E2, and include or exclude sequences that are located in that region but do not cover it completely. Retrieved sequences and the associated annotation can be downloaded as an alignment which will usually be codon-aligned, so that it can be translated immediately, or as translated amino acids in any reading frame. The retrieved background information can also be downloaded as a tab-delimited file.
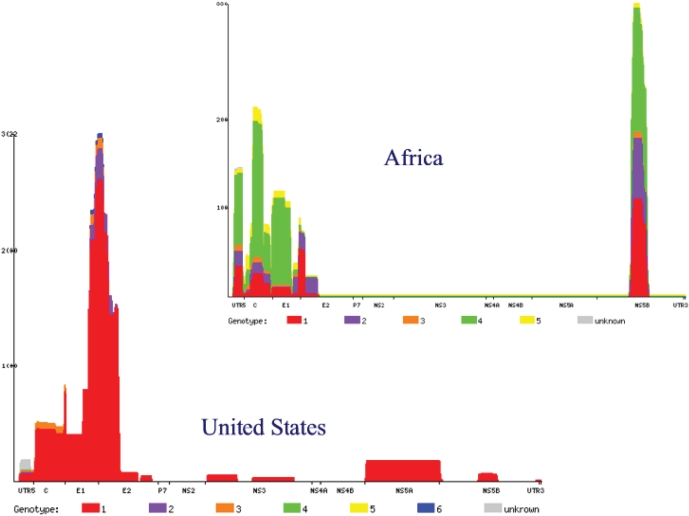
Figure 1.
Histograms showing the number, genotype and genomic region for two sets of sequences: all those sampled in Africa, and all those sampled in the United States of America. Please note that the scales of the vertical axes are different. It can immediately be seen that there are many more sequences from the USA; that they are mostly of genotype 1 whereas African sequences are of all genotypes; and that the best region for comparative analysis right now (because most background sequences would be available) in Africa would be NS5B or Core/E1, while for the US it would be E1/E2.
The search interface includes two other important features. First, it allows users to automatically align their own sequences with any database sequences that cover the same region of the genome, and to download the resulting alignment. Second, any alignment that has been retrieved can be used to build a tree immediately, as sequences are internally aligned. The tree-making module also gives the option of adding the appropriate genotype reference sequences to the alignment, so the sequences can be genotyped very easily.
Some fields in the HCV database, such as the patients HLA type, are still very lightly populated, comparing poorly with comparable fields in the HIV database which is maintained by the same group. This is often because the information is not made available by the authors. We hope that the fact that these data are now stored centrally and made easily available will encourage authors to provide more comprehensive information, as has indeed happened for HIV.
TOOLS PROVIDED ON THE WEBSITE
The list here is very partial; a more elaborate review of the possibilities of the HCV database can be found in Ref. (10). The website provides manually optimized HCV alignments that do not contain any closely related sequences, as well as reference alignments, which contain 3–4 representatives of each available genotype and subtype and can be used as background sequences in phylogenetic trees. For all genes and proteins, a graphical overview is presented of their overall and synonymous/nonsynonymous variation (using sliding window analysis) and the cumulative variation of each gene/protein. Also provided is an alignment of flavivirus complete genomes, along with some results of divergence analysis of these viruses.
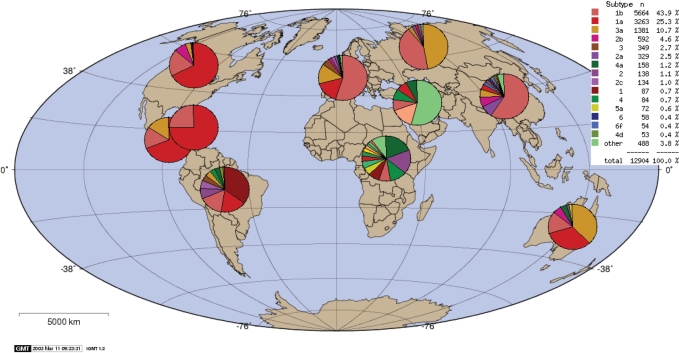
Figure 2 shows output from the ‘Geography tool’, which can be used to plot frequencies of the different genotypes stored in the database as a function of their geographical origin. This tool can be very useful to get a general idea of which genotypes have been found in which countries, as well as the density of sampling in different regions of the world.
Figure 2.
World map showing the geographical distribution of sequences with different genotypes in the HCV database. The map was generated using the geography tool, available on the HCV website.
Other available tools for common types of sequence analysis, tailored to HCV sequence data, include:
‘Syn-Nonsyn’ calculates, analyzes and builds trees from the numbers of silent (ds) and nonsilent (dn) mutations in a codon alignment (11); ‘Glycosite’ tallies and plots N-linked glycosylation sites (12); ‘Entropy’ plots the variability of each portion in an alignment, and can test differences between two alignments.
In addition, there are interfaces to several public domain programs:
‘Treemaker’ generates Neighbor-joining trees. It is a user-friendly interface to the DNAdist/Neighbor/Drawtree suite from the PHYLIP package (13). The interface works around several common problems: it gapstrips the alignments before feeding them into DNAdist, it preserves sequence names longer than 10 characters, and removes negative numbers in the calculated distance matrix that can cause the programs to crash. ‘BLAST’ (14) searches the HCV database for nucleotide sequences that are most similar to the user's query sequence. The search can be limited to sequences that have a valid genotype.
‘PCOORD’ (15) offers a principal coordinate analysis, a data-reduction technique similar to principal components analysis to identify co-varying positions in groups of sequences.
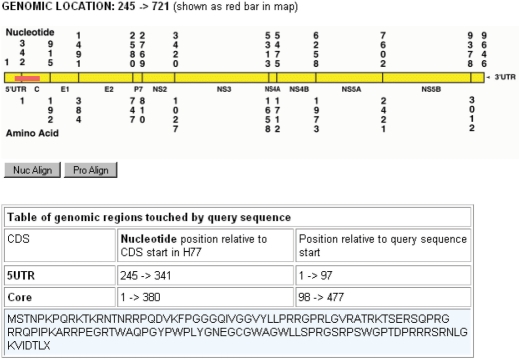
Finally, a large number of tools are made available for manipulating or summarizing sequences. Some examples: ‘FindModel’ is a variation of Modeltest (16) a procedure that finds the evolutionary model that is most suitable for a given dataset and lists its parameters. ‘Gene Cutter’ is an HCV-specific tool that finds defined genes in a nucleotide input sequence, generates the appropriate amino acid translation, and is able to codon-align a set of sequences, which can then be downloaded. ‘Consensus’, a versatile tool to create consensus sequences of groups of sequences that can be modified by a large number of parameters. ‘Branchlength’ calculates the length of the branches in a tree between two nodes, or from a chosen node to the endnotes below it. It can also be used to re-root and plot a tree. ‘Sequence Locator’ is a program that finds the coordinates of an input sequence relative to the reference strain H77 (Figure 3). This program can be used as a means to standardize primer and epitope numbering, and quickly shows the user the location of an unknown HCV sequence fragment. It provides the amino acid translation in the correct frame if a nucleotide sequence was submitted, and aligns the amino acid sequence against the HCV-H77 nucleotide sequence if the input is an amino acid sequence. Sequence locator can also be used for reverse-complement sequences. ‘PeptGen’ is intended to help immunologists rationally design overlapping peptide sets to probe the immune response, taking into account forbidden N- and C-terminal amino acids and desired peptide length. ‘Primalign’ and ‘Epilign’ automatically align a primer or epitope to the HCV complete genome alignment. The interface returns the coordinates (H77 numbering) and an alignment of the fragment to all sequences in the whole genome alignment.
Figure 3.
Output of the Sequence Locator tool, which finds the location of a sequence fragment of any length. The graphic shows where the user's input sequence is located; the table beneath it shows the amino acid translation and coordinates of all of the genes included in the fragment.
OTHER FUNCTIONS OF THE HCV DATABASE
The database staff also tries to facilitate HCV sequence research and analysis in other ways. For example, the database took the initiative and secured NIH-DMID funding to try to fill the patchy record of HCV complete genomes of all genotypes. Currently, complete genomes are available for only 31 of the 71 officially established geno/subtype variants. The isolation and sequencing is being done by various groups, and the database further supports the effort by publicizing and finding potential sample sources. The database group also supported an initiative to standardize the HCV nomenclature (17) and the numbering of genes and region in the HCV genome (10).
FUTURE ENHANCEMENTS
Several new tools are close to being made public. The first one is a tool named Phyloplace. It will help users to both decide which genotype and subtype their sequence resembles, and if it is different from all, whether it is different enough to be called a new subtype or even a new genotype. Results indicate that the phylogenetic placement can be done and recombinants can be detected with reasonable accuracy (manuscript in preparation). Recombination in HCV, while known, so far seems to be a relatively rare phenomenon. A second tool, Distplot, will help users calculate, analyze and generate graphs of matrices of pairwise distances. It will take a sequence alignment and (when applicable) grouping information, calculate various distance measures, provide summary statistics and significance tests and plot the distances relative to each other or to one sequence or group (e.g. the first sample in a time series).
ACKNOWLEDGEMENTS
We thank Bette Korber and our colleagues at the HIV databases for their generous help and support. We also gratefully acknowledge the help of the members of the Editorial Board for many suggestions for improvement. The HCV database is funded by the Division of Microbiology and Infectious Diseases of the National Institute of Allergies and Infectious Diseases (NIAID).
Conflict of interest statement. None declared.
REFERENCES
- 1.Alter MJ, Margolis HS, Krawczynski K, Judson FN, Mares A, Alexander WJ, Hu PY, Miller JK, Gerber MA, et al. The natural history of community-acquired hepatitis C in the United States. The Sentinel Counties Chronic non-A, non-B Hepatitis Study Team. N. Engl. J. Med. 1992;327:1899–1905. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- 2.Hoofnagle JH. Hepatitis C: the clinical spectrum of disease. Hepatology. 1997;26:15S–20S. doi: 10.1002/hep.510260703. [DOI] [PubMed] [Google Scholar]
- 3.Krahn M, Wong JB, Heathcote J, Scully L, Seeff L. Estimating the prognosis of hepatitis C patients infected by transfusion in Canada between 1986 and 1990. Med. Decis. Making. 2004;24:20–29. doi: 10.1177/0272989X03261568. [DOI] [PubMed] [Google Scholar]
- 4.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 5.Vrielink H, Zaaijer HL, Reesink HW, van der Poel CL, Cuypers HT, Lelie PN. Sensitivity and specificity of three third-generation anti-hepatitis C virus ELISAs. Vox Sang. 1995;69:14–17. doi: 10.1111/j.1423-0410.1995.tb00341.x. [DOI] [PubMed] [Google Scholar]
- 6.CDC. Transmission of hepatitis B and C viruses in outpatient settings – New York, Oklahoma, and Nebraska, 2000-2002. MMWR Morb. Mortal. Wkly Rep. 2003;52:901–906. [PubMed] [Google Scholar]
- 7.Simmonds P. Viral heterogeneity of the hepatitis C virus. J. Hepatol. 1999;31(Suppl. 1):54–60. doi: 10.1016/s0168-8278(99)80375-4. [DOI] [PubMed] [Google Scholar]
- 8.Farci P, Purcell RH. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin. Liver Dis. 2000;20:103–126. [PubMed] [Google Scholar]
- 9.Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 10.Kuiken C, Combet C, Bukh J, Shin-I T, Deleage G, Mizokami M, Richardson R, Sablon E, Yusim K, et al. A comprehensive system for consistent numbering of HCV sequences, proteins and epitopes. Hepatology. 2006;44:1355–1361. doi: 10.1002/hep.21377. [DOI] [PubMed] [Google Scholar]
- 11.Korber B. In: Computational Analysis of HIV Molecular Sequences. Rodrigo AG, Learn GH, editors. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. [Google Scholar]
- 12.Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes, and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein J. V3.5. 3.52c edn. Seattle, WA: University of Washington; 1984. PHYLIP: Phylogeny Inference Package. [Google Scholar]
- 14.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Higgins DG. Sequence ordinations: a multivariate analysis approach to analysing large sequence data sets. Comput. Appl. Biosci. 1992;8:15–22. doi: 10.1093/bioinformatics/8.1.15. [DOI] [PubMed] [Google Scholar]
- 16.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 17.Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]