Abstract
Medaka (Oryzias latipes) is a small egg-laying freshwater teleost native to East Asia that has become an excellent model system for developmental genetics and evolutionary biology. The draft medaka genome sequence (700 Mb) was reported in June 2007, and its substantial genomic resources have been opened to the public through the University of Tokyo Genome Browser Medaka (UTGB/medaka) database. This database provides basic genomic information, such as predicted genes, expressed sequence tags (ESTs), guanine/cytosine (GC) content, repeats and comparative genomics, as well as unique data resources including (i) 2473 genetic markers and experimentally confirmed PCR primers that amplify these markers, (ii) 142 414 bacterial artificial chromosome (BAC) and 217 344 fosmid end sequences that amount to 15.0- and 11.1-fold clone coverage of the entire genome, respectively, and were used for draft genome assembly, (iii) 16 519 460 single nucleotide polymorphisms (SNPs), and 2 859 905 insertions/deletions detected between two medaka inbred strain genomes and (iv) 841 235 5′-end serial analyses of gene-expression (SAGE) tags that identified 344 266 transcription start sites on the genome. UTGB/medaka is available at: http://medaka.utgenome.org/
INTRODUCTION
Teleosts comprise more than half of all vertebrate species and have adapted to a variety of marine and freshwater habitats (1). For the past two decades, smaller teleosts, such as zebrafish and medaka, have been used to study various aspects of basic biology because of the power of forward genetics and the availability of elegant embryo manipulation techniques. Medaka has frequently been used for carcinogenesis studies and for testing endocrine disruptors in ecotoxicology analysis (2). Furthermore, medaka research has a long, outstanding history in the field of sex determination: it facilitated the first demonstration of Y-linked inheritance in any species (3), the first successful sex-reversal in vertebrates (4) and the identification of the male-determining gene, DMY, the first non-mammalian functional equivalent of SRY (5). Highly polymorphic and healthy inbred strains of medaka have been used for both mutagenesis screening and genetic mapping. Furthermore, medaka research has recently been boosted by the successful generation of hundreds of developmental mutants (6). Significantly, some of these mutants have unique phenotypes that are not covered by zebrafish mutants, probably due to different gene repertoires. These features make medaka an attractive system for developmental genetics. To afford users quick access to medaka developmental biology resources, UTGB/medaka includes various unique datasets, as well as sophisticated genomics tools. While general purpose genome browsers, Ensembl and UCSC (7,8), courteously provide the medaka genome and its fundamental features (e.g. predicted genes, ESTs, GC content, repeats and comparative genomics), most of genomic resources described in this article are obtainable only from UTGB/medaka because these resources are tailored to specific needs in medaka biology and are designed to facilitate both designing further biological experiments and obtaining libraries of interest.
DATA RESOURCES
Genome sequences
The genome of Hd-rR, an inbred medaka strain, was assembled from 13.8 million reads that were obtained from the whole genome shotgun plasmid, fosmid and bacterial artificial chromosome (BAC) libraries (9). The total size of the assembled contigs was 700.4 megabases (Mb), although previous estimates range from 851 to 1080 Mb, according to the measurement of haploid weight (10–13). This discrepancy was an inevitable result of selecting the whole-genome shotgun sequencing strategy, which is cost efficient but suffers from inherent technical limitations. For example, highly homologous repetitive elements are likely to be grouped into small contigs, and are rarely anchored onto unique positions in the genome owing to their redundancy (9). Therefore, the assembled contigs largely represent the medaka genome but may fail to include highly repetitive elements. Of the 700.4 Mb in the sequenced genome, 50% of nucleotides are covered in scaffolds (or contigs) of length > 1.41 Mb (9.8 kb) that are called N50 values. This contiguity is sufficient to characterize the genomic structures of genes.
The medaka genome sequence data have been released to the public four times to meet urgent requests from the medaka research community. Four versions named 200406, 200506, version 0.9 and version 1.0 have been created to provide users with timely information. The former two versions had shorter scaffolds that were not anchored on the medaka chromosomes because they were built in 2004 and 2005, before genetics markers were available. Versions 0.9 and 1.0 were created in 2006, when comprehensive genetic markers were available, so that about 90% of their scaffolds and ultracontigs were located on the 24 medaka chromosomes. Versions 0.9 and 1.0 were built from the identical contigs and scaffolds, but the assembly of version 1.0 is longer than that of version 0.9 because more genetic markers could be used to generate version 1.0. Version 0.9 is left open to the public because most of the data analysis in the medaka genome paper (9) was based on version 0.9. In these two versions, two scaffolds linked by a single BAC are connected into one ultracontig if it is consistent with genetic markers. The N50 value of ultracontigs in version 1.0 amounted to 5.1 Mb, excluding gaps, and therefore, the great continuity of ultracontigs promises to accelerate the task of positional cloning with an ample number of confirmed genetic markers in our database.
BAC/Fosmid end sequences
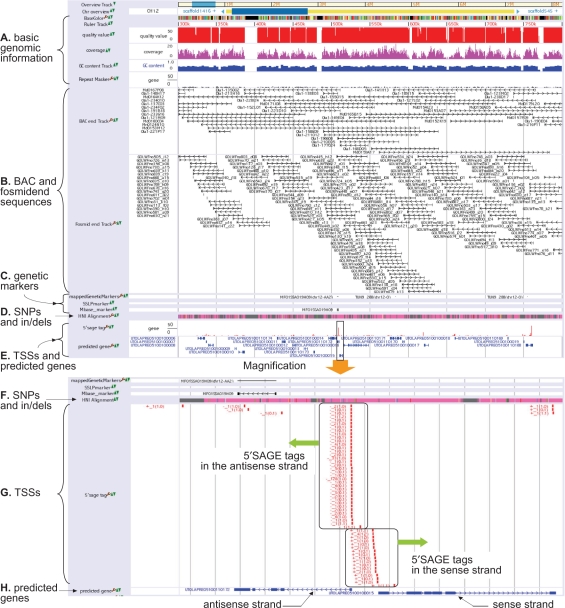
Another principal data resource in our database is the fosmid and BAC end sequences anchored on the medaka chromosomes. Sequencing a gene of interest makes it possible to map the uncovered sequence of the gene onto the genome, but the aligned region may still contain some gaps to be finalized because of the aforementioned limitations in the whole-genome shotgun sequencing method. The availability of BAC or fosmid clones that cover the genomic region of interest is indispensable to the task of filling gaps and finalizing the region. To assist this process, our genome browser displays the positions of end sequences of BAC and fosmid clones surrounding the genomic region of interest (see Figure 1B). Actually, sufficient fosmid and BAC clones have been collected for sequencing the medaka genome; namely, 217 344 fosmid end sequences and 142 414 BAC end sequences have been mapped on the genome. To be more precise, two types of fosmid library are of average size 35.5 and 37.5 kb, and three types of BAC library are of average size 135, 180 and 210 kb. The respective clone coverage of fosmids and BACs are 11.1 and 15.0 times of the medaka genome, demonstrating that sufficient fosmid and BAC libraries are available for completing a specific region.
Figure 1.
Snapshot of UTGB/medaka. (A) Basic genomic information such as bases, quality values and sequence coverages of individual bases, GC content and repeats. (B) Positions of BAC and fosmid end sequences. Right (or left) arrows mean that the BAC/fosmid clone is in the sense (antisense) strand. (C) Single nucleotide polymorphism (SNP), single sequence length polymorphism (SSLP) and restriction fragment length polymorphism (RFLP) markers. (D) SNPs and insertions/deletions between medaka inbred strains Hd-rR and HNI. Details are shown in the magnification view (Figure 1F). (E) In the upper row, the height of a red bar presents the number of 5′SAGE tags observed at the position, while the lower row displays genes predicted from TSSs identified by 5′SAGE tags. The range enclosed in the box is magnified and shown in the lower portion. (F) SNPs and indels. Pink indicates matched bases, while mismatches are highlighted by green, yellow, blue and red that represent A, C, G and T, respectively, in the HNI genome. Cyan indicates the two possibilities, deletions from the HNI genome or insertions into the Hd-rR genome, though the settlement requires comparison with the genome of an outgroup of Hd-rR and HNI. Gray means the failure of the alignment because of low quality bases in either of the two genomes or lack of HNI reads in the region. (G) The positions of individual 5′SAGE tags are illustrated by red boxes. The label associated with each 5′SAGE tag is of the form S_F (L0, L1), where S indicates whether the tag is derived from the sense (+) or the antisense strand (−) of the position, F is the frequency of occurrences of the tag in the collection from a mixture of cDNA, and L0 (L1, resp.) means the number of loci where the tag is aligned to the genome with no mismatch (with one mismatch). Tags are aligned to unique positions if L0 = 1, or L0 = 0 and L1 = 1. For example, a tag labeled with +_4(1,2) is observed four times in the tag collection, it is aligned uniquely to the plus strand of the position without mismatch, and it is also aligned to two other locations with one mismatch. A tag with −_1(0,1) occurs once in the collection and fails to map to the genome with no mismatch but maps uniquely to the minus strand of the position with one mismatch. The figure displays a large number of 5′SAGE tags are observed in both strands. (H) The structures of two predicted genes are shown. Boxes represent protein-coding exons. Right (or left) arrows indicate that the gene is encoded in the sense (antisense) strand.
Genetic markers
In addition to the genome of the Hd-rR medaka inbred strain, the genome of another inbred strain HNI (14,15) was also sequenced to produce the draft 648-Mb HNI genome. Inbred strains Hd-rR and HNI originated in the southern and northern Japanese populations, respectively. They can mate and produce healthy offspring, although they are estimated to have diverged about 4 million years ago, and their genome sequences have diverged by ∼3.42%. The alignment of the two medaka genomes identified about 16.4 million single nucleotide polymorphisms (SNPs), from which 2401 SNPs were selected and mapped genetically onto medaka chromosomes using a backcross panel between these two inbred strains. These genetic SNP markers, together with 140 single sequence length polymorphism (SSLP) and restriction fragment length polymorphism (RFLP) markers, were used to anchor scaffolds on chromosomes, to construct the medaka chromosome map. These confirmed markers should be useful in isolating responsible genes of interest by positional cloning. To enable users to use the markers, our database contains PCR primers for 2473 markers, with the genetic distances between the markers, and their locations on the chromosomes (Figure 1C).
SNPs and insertions/deletions
The alignment of the Hd-rR and HNI genomes revealed 16 519 460 SNPs and 2 859 905 insertions/deletions (indels), and the locations of individual SNPs and indels are given in the web database (Figure 1D and F). This information constitutes an excellent resource for exploring a variety of further studies. One typical use is to design PCR primers for SNP markers concentrated in a particular genomic region of interest, to boost positional cloning. Such SNP markers would also be attractive for characterizing the genetic divergence among various inbred and wild-type medaka strains found in Asia. For example, the sequences surrounding a number of SNP markers were collected from four inbred and eight wild-type strains, and the multiple alignment of these sequences generated a phylogenetic tree that was consistent with the regional separation of these strains and confirmed the recent expansion of the northern Japanese population (9). Another intriguing direction would be to analyze whether a specific gene and its regulatory region are evolving slowing or rapidly, by examining the two inbred strains. For instance, genes in the reproduction and sex-related categories were shown to be evolving slowlyly relative to the hominid lineage, presumably because these two inbred strains have not speciated (9). Overall, the set of SNPs and indels reflects a comprehensive view of genetic divergence between the two medaka inbred strains, which is useful in putting forward medaka genetics.
Transcription start sites, predicted genes and gene expression
Given the limited availability of expressed gene tags (EST) and full-length enriched cDNA sequences, a considerable amount of 5′-end serial analyses of gene expression (5′SAGE) data (16), the 19–20 5′-end bases of mRNAs, have been collected, to detect transcription start sites (TSSs) and subsequently protein-coding regions were predicted from the TSSs by using GENSCAN (17). In all, 1 186 742 5′SAGE tags were collected from a mixture of cDNA from 0- to 7-day-old medaka embryos and adult body tissues. Of these, 841 235 (70.9%) were aligned to unique positions in the medaka draft genome, but most of them were duplicates and were expressed from 344 266 transcription start sites. Stated another way, multiple tags are often derived from one locus, and the frequency approximates the expression level of the gene encoded at that locus (Figure 1E and G). From these TSSs, 20 141 genes were predicted (Figure 1E and H), and individual predicted genes were supported by the evidence of some 5′ SAGE tags, which was effective in reducing the false-positive ratio of predicted genes. These evidence-based predicted medaka genes were compared with human genes comprehensively, and about 57.7% of the predicted medaka genes have human orthologs, and 21.6% of the genes constitute medaka-human reciprocally best 1:1 ortholog pairs (9), indicating that medaka could serve as a model system for humans.
SEARCHING THE GENOME
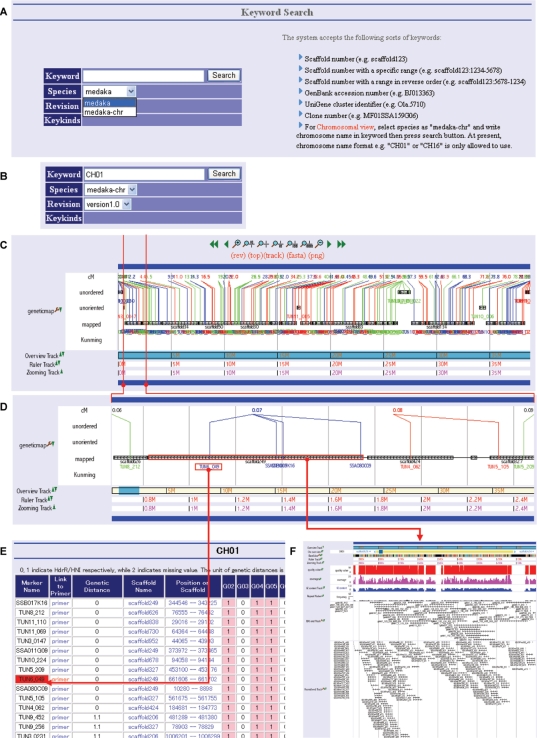
In UTGB/medaka, biological data are organized along the medaka genome sequence. One can approach a genomic region of interest in three different ways. The first is the typical method that searches for proper keywords, such as scaffold names, gene names and EST accession numbers. The second is to input a sequence of interest, such as a cloned and sequenced gene, into the online mapping window, and subsequently the system returns the list of location candidates of the given sequence with their alignments to the genome and displays the selected location in the main window. The last is to display an overview of all genetic markers on individual chromosomes that are useful in boosting the positional cloning of genes responsible for a specific phenotype. Our database provides a high-resolution mapping of confirmed genetic markers as well as genomic sequences on which individual genetic markers are located. As shown in Figure 1, genomic sequences are associated with a variety of information, e.g. predicted genes, 5′SAGE tags, and ESTs. In the meanwhile, Figure 2 illustrates how to browse the list of genetic markers along the chromosome of interest. The maps of genetic markers shown in Figure 2C and D facilitate chromosome walking towards the locus that is likely to contain responsible genes, and clicking the scaffold on the locus presents precise information, e.g. candidate genes, as shown in Figure 2F.
Figure 2.
Genetic markers. (A) To display genetic markers on the chromosome of interest, start with the top-level ‘Keyword Search’ view by selecting species as ‘medaka-chr.’ (B) Specify the chromosome number by inputting “CH” followed by the number, e.g. ‘CH01’ or ‘CH16’. (C) Clicking the ‘Search’ button presents all genetic markers horizontally along the chromosome. The distance of a genetic marker from the head of the chromosome is shown in terms of centimorgan (cM). Ultracontigs (series of scaffolds) are categorized into three groups, mapped, unoriented and unordered. Mapped ultracontigs are anchored on the chromosome by multiple genetic markers among which at least one recombination is observed. Unoriented ultracontigs are associated to the specific position on the genetic map, but their directions are unknown because no recombination is observed in the ultracontigs. A cluster of ultracontigs is unordered if it is located on the genetic map but neither the order in the cluster nor the orientation is known because no recombination is observed in the cluster. (D) An enlarged view of the window bounded by two vertical, red lines in Figure 2C. The ultracontig illustrated in the figure has four scaffolds linked by horizontal black lines that represent BAC end pairs. The four blue lines connected to one of the scaffolds from the node labeled with 0.07 cM indicate genetic markers among which no recombination is observed. (E) Clicking the leftmost, blue genetic marker name presents the list of markers surrounding the selected marker. (F) Selecting the scaffold enclosed in the red box displays the view with a variety of precise information associated with the scaffold, which is similar to Figure 1.
DATA DOWNLOAD
All of our sequence and annotated data are available for download in various formats described in http://download.utgenome.org/pub/. The latest version 1.0 genomic sequences are registered in DDBJ/GenBank/EMBL and have been released as accession numbers BAAF04000001-BAAF04134429 (contigs), DF083412-DF090103 (scaffolds), DF090104-DF090315 (ultracontigs) and DG000001-DG000024 (chromosomes). The HNI assembly version 1.0 is registered as BAAE01000001-BAAE01346141 (contigs) and DF000001-DF038235 (scaffolds). In addition, the 5′SAGE tag data are registered as accession numbers, ACAAA0000001-ACAAA0356693. BAC end reads are available as accession numbers DE248528-DE283656 (collected at National Institute of Informatics) and DE042646-DE143283 (Keio University) (18).
CONCLUSION AND FUTURE PLAN
To fully exploit the power of the medaka genome for medaka and developmental biology, UTGB/medaka is equipped with data information on considerable biological resources and advanced genomic tools. To support further experiments, we plan to add a number of other useful data, e.g. RFLP primers and phenotype information of well-known mutants.
ACKNOWLEDGEMENTS
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas ‘Genome’ from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), and the Japan Science and Technology Corporation (JST). We thank the Human Genome Center, University of Tokyo for computational time, and the National BioResource Project of MEXT for supplying the medaka strain and for other support. Funding to pay the Open Access publication charges for this article was provided by JST.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nelson JS. Fishes of the World. 3rd. New York: Wiley; 1994. [Google Scholar]
- 2.Patyna PJ, Davi RA, Parkerton TF, Brown RP, Cooper KR. A proposed multigeneration protocol for Japanese medaka (Oryzias latipes) to evaluate effects of endocrine disruptors. Sci. Total Environ. 1999;233:211–220. doi: 10.1016/s0048-9697(99)00227-2. [DOI] [PubMed] [Google Scholar]
- 3.Aida T. On the inheritance of color in a fresh-water fish, Aplocheilus latipes Temminck and Schlegel, with special reference to sex-linked inheritance. Genetics. 1921;6:554–573. doi: 10.1093/genetics/6.6.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto T. Artificially induced sex-reversal in genotypic males of the Medaka (Oryzias latipes) J. Exp. Zool. 1953;123:571–594. [Google Scholar]
- 5.Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey CE, Shibata N, Asakawa S, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- 6.Furutani-Seiki M, Sasado T, Morinaga C, Suwa H, Niwa K, Yoda H, Deguchi T, Hirose Y, Yasuoka A, et al. A systematic genome-wide screen for mutations affecting organogenesis in Medaka, Oryzias latipes. Mech. Dev. 2004;121:647–658. doi: 10.1016/j.mod.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 7.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- 10.Hinegardner R, Rosen DE. Cellular DNA content and the evolution of teleostean fishes. Am. Nat. 1972;106:621–644. [Google Scholar]
- 11.Uwa H, Iwata A. Karyotype and cellular DNA content of Oryzias javanicus (Oryziatidae, Pisces) Chromosome Inf. Service. 1981;31:24–26. [Google Scholar]
- 12.Uwa H. Karyotype evolution and geographical distribution in the ricefish, genus Oryzias (Oryziidae) Indo-Pacific Fish Biol. 1986:867–876. [Google Scholar]
- 13.Lamatsch DK, Steinlein C, Schmid M, Schartl M. Noninvasive determination of genome size and ploidy level in fishes by flow cytometry: detection of triploid Poecilia formosa. Cytometry. 2000;39:91–95. doi: 10.1002/(sici)1097-0320(20000201)39:2<91::aid-cyto1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 14.Naruse K, Hori H, Shimizu N, Kohara Y, Takeda H. Medaka genomics: a bridge between mutant phenotype and gene function. Mech. Dev. 2004;121:619–628. doi: 10.1016/j.mod.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Wittbrodt J, Shima A, Schartl M. Medaka–a model organism from the far East. Nat. Rev. Genet. 2002;3:53–64. doi: 10.1038/nrg704. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto S, Suzuki Y, Kasai Y, Morohoshi K, Yamada T, Sese J, Morishita S, Sugano S, Matsushima K. 5′-end SAGE for the analysis of transcriptional start sites. Nat. Biotechnol. 2004;22:1146–1149. doi: 10.1038/nbt998. [DOI] [PubMed] [Google Scholar]
- 17.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki T, Shimizu A, Ishikawa SK, Imai S, Asakawa S, Murayama Y, Khorasani MZ, Mitani H, Furutani-Seiki M, et al. The DNA sequence of medaka chromosome LG22. Genomics. 2007;89:124–133. doi: 10.1016/j.ygeno.2006.09.003. [DOI] [PubMed] [Google Scholar]