Abstract
AAindex is a database of numerical indices representing various physicochemical and biochemical properties of amino acids and pairs of amino acids. We have added a collection of protein contact potentials to the AAindex as a new section. Accordingly AAindex consists of three sections now: AAindex1 for the amino acid index of 20 numerical values, AAindex2 for the amino acid substitution matrix and AAindex3 for the statistical protein contact potentials. All data are derived from published literature. The database can be accessed through the DBGET/LinkDB system at GenomeNet (http://www.genome.jp/dbget-bin/www_bfind?aaindex) or downloaded by anonymous FTP (ftp://ftp.genome.jp/pub/db/community/aaindex/).
INTRODUCTION
Protein structures and functions are defined by the combinations of physicochemical and biochemical properties of 20 naturally occurring amino acids that are the building-blocks of proteins. A wide variety of properties of amino acids have been investigated through a large number of experiments and theoretical studies. Each of these amino acid properties that can be represented by a set of 20 numerical values is referred to as an amino acid index. Nakai et al. (1) collected 222 amino acid indices from published literature and investigated the relationships among them using hierarchical cluster analysis. They also released the amino acid indices as an online database. In 1996, Tomii and Kanehisa (2) further collected amino acid indices to enrich the database. Additionally, they also collected 42 amino acid substitution matrices from the literature and released the collection as AAindex2. The AAindex database is continuously updated by the present authors (3,4).
AAindex has been used in wide-ranging bioinformatics research on protein sequences, such as predicting protein subcellular localization (5), immunogenicity of MHC class I binding peptides (6), protein SUMO modification site (7) and coordinated substitutions in multiple alignments of protein sequences (8). Furthermore, there is a derivative database of AAindex (UMBC AAindex Database: http://www.evolvingcode.net:8080/aaindex/) and a web tool for visualizing relationships among AAindex entries (9). Given the examples cited here, AAindex has become a useful resource in bioinformatics.
In 2005, Pokarowski et al. (10) compared 29 published matrices of protein pairwise contact potentials, i.e. energy functions that are obtained from statistical analysis of protein structures (10). These potentials have long been used to predict protein structures in silico. Pokarowski and coworkers elucidated that each of the contact potentials is similar to one of two popular matrices derived by Miyazawa and Jernigan (11). Recently, working on 29 mostly new amino acid substitution matrices and 5 contact potentials, the same team (12) obtained segregation of substitution matrices similar to Tomii and Kanehisa (2). Moreover, they found intermediate links between substitution matrices and contact potentials—matrices and potentials that exhibit mutual correlations of at least 0.8. In both works (10,12), Pokarowski and coworkers approximated matrices by simple functions of amino acid indices, which allow us to comprehend better the exchangeability of amino acids as well as the residue–residue interactions in proteins. These relations between substitution matrices, contact potentials and amino acid indices provide motivation to extend the AAindex database. In the present work, we have compiled the data collected in the study on contact potentials (10) as a new section of AAindex database, named AAindex3. As a result we believe that the AAindex has increased its utility in the bioinformatics study of proteins. In this paper we report the current status of the three sections of AAindex.
THE CURRENT DATABASE
The AAindex is released approximately annually. The latest version is the 9.0 release.
The AAindex database is a flat file database that consists of three sections: AAindex1 for the amino acid indices, AAindex2 for the amino acid substitution matrices and AAindex3 for the amino acid contact potentials. The contents of the three sections are as follows.
AAindex1
The AAIndex1 currently contains 544 amino acid indices. Each entry consists of an accession number, a short description of the index, the reference information and the numerical values for the properties of 20 amino acids.
We have provided a link to the corresponding PubMed entries of each AAindex entry, instead of a link to the LitDB literature database (13) that we originally used. In addition, each entry contains cross-links to other entries with an absolute value for the correlation coefficient of 0.8 or larger. The links enable the users to identify a set of entries describing similar properties. In some instances the values are not reported for all 20 amino acids.
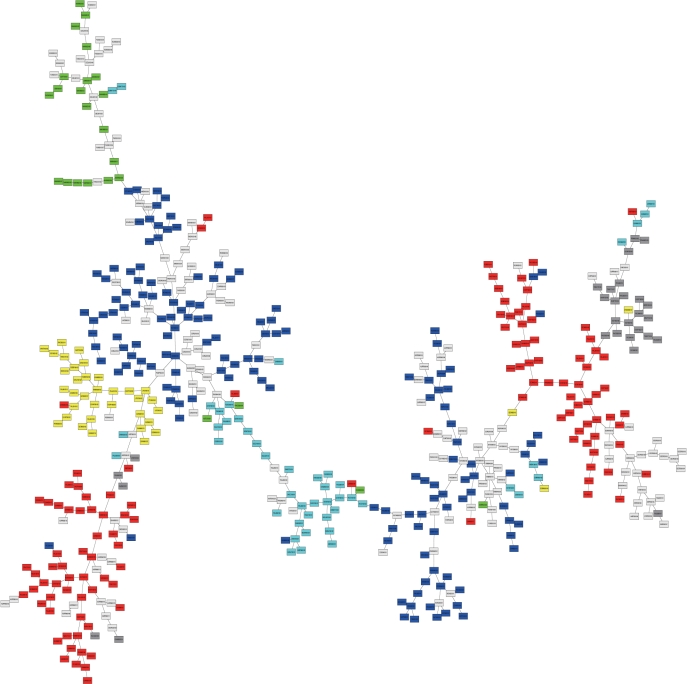
To represent an overview of the relationships among current amino acids indices, we constructed the minimum spanning tree of amino acid indices by the procedure described by Tomii et al. (2) (Figure 1). In Figure 1, each rectangle represents an index. The colored rectangles are the 402 indices classified in six groups defined by Tomii and coworkers. The indices belonging to the Tomii's classification are still grouped into clusters. Newly added indices are distributed evenly across the tree. That is, the indices for various kinds of properties have been added to the AAindex.
Figure 1.
The minimum spanning tree of the amino acid indices stored in the AAindex1 release 9.0. Each rectangle is an amino acid index. Colored nodes represent the indices classified by Tomii et al. (2) Red: alpha and turn propensities, Yellow: beta propensity, Green: composition, Blue: hydrophobicity, Cyan: physicochemical properties, Gray: other properties. White: the indices added to the AAindex after the release 3.0 by Tomii et al. (2).
AAindex2
The AAindex2 currently contains 94 amino acid substitution matrices: 67 symmetric matrices and 27 non-symmetric matrices. The format of the entry is almost the same as that of AAindex1 except that it contains 210 numerical values (20 diagonal and 20 × 19/2 off-diagonal elements) for a symmetric matrix and 400 or more numerical values for a non-symmetric matrix (some matrices include a gap or distinguish two states of cysteine). In the previous release, each symmetric matrix, which is triangular in shape, was folded into a 10 × 21 table for the purpose of saving space, and columns were separated by space characters. In the present release, symmetric matrices are not folded and delimiter of columns has been changed into a tab character easier parsing of the entry.
AAindex3
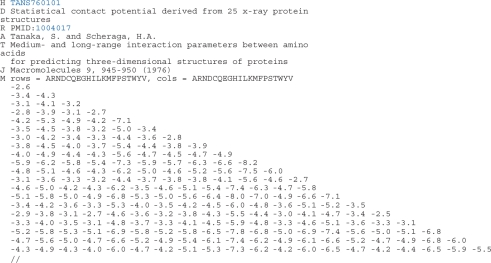
The AAindex3 section currently contains 47 amino acid contact potential matrices: 44 symmetric matrices and 3 non-symmetric matrices. The format of the entry is almost the same as that of AAindex2. A sample entry of the AAindex3 is shown in Figure 2.
Figure 2.
An example of database entry in the AAindex3. Each record of an entry is identified by the one-letter codes: H, accession number; D, definition of the entry; R, PMID identifier; A, author(s); T, title of the journal article; J, journal citation information; M, actual data in the specified order.
AVAILABILITY
The AAindex database can be retrieved through the DBGET/LinkDB system (14) of the Japanese GenomeNet service (15) at http://www.genome.jp/dbget-bin/www_bfind?aaindex.
The DBGET/LinkDB system integrates most of the major molecular biology databases and is especially suited for using hyperlinks to related entries within the AAindex database as well as to the other databases. Alternatively, the entries database may be copied and used locally. The URL for anonymous FTP is: ftp://ftp.genome.jp/pub/db/community/aaindex/
BioRuby that is a bioinformatics library of Ruby programming language has provided the useful functions to handle the AAindex database (http://bioruby.org/). EMBOSS (16) has provided a program to extract the index data from the AAindex entry.
Users are requested to cite this article when making use of the AAindex database.
ACKNOWLEDGEMENTS
We thank Drs Kenta Nakai and Kentaro Tomii for the initial developments of the AAindex database. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology, and the Japan Science and Technology Agency. We thank Ms Mansi Srivastava and Dr Takeshi Kawashima for critical reading of our manuscript. The computational resources were provided by the Bioinformatics Center, Institute for Chemical Research, Kyoto University and the Super Computer System, Human Genome Center, Institute of Medical Science, University of Tokyo. Funding to pay the Open Access publication charges for this article was provided by the University of Tokyo.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nakai K, Kidera A, Kanehisa M. Cluster analysis of amino acid indices for prediction of protein structure and function. Protein Eng. 1988;2:93–100. doi: 10.1093/protein/2.2.93. [DOI] [PubMed] [Google Scholar]
- 2.Tomii K, Kanehisa M. Analysis of amino acid indices and mutation matrices for sequence comparison and structure prediction of proteins. Protein Eng. 1996;9:27–36. doi: 10.1093/protein/9.1.27. [DOI] [PubMed] [Google Scholar]
- 3.Kawashima S, Ogata H, Kanehisa M. AAindex: amino acid index database. Nucleic Acids Res. 1999;27:368–369. doi: 10.1093/nar/27.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawashima S, Kanehisa M. AAindex: amino acid index database. Nucleic Acids Res. 2000;28:374. doi: 10.1093/nar/28.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarda D, Chua GH, Li K-B, Krishnan A. pSLIP: SVM based protein subcellular localization prediction using multiple physicochemical properties. BMC Bioinformatics. 2005;6:152. doi: 10.1186/1471-2105-6-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tung C-W, Ho S-Y. POPI: predicting immunogenicity of MHC class I binding peptides by mining informative physicochemical properties. Bioinformatics. 2007;23:942–949. doi: 10.1093/bioinformatics/btm061. [DOI] [PubMed] [Google Scholar]
- 7.Liu B, Li S, Wang Y., c, Lu L, Li Y, Cai Y. Predicting the protein SUMO modification sites based on properties sequential forward selection (PSFS) Biochem. Biophys. Res. Comm. 2007;358:136–139. doi: 10.1016/j.bbrc.2007.04.097. [DOI] [PubMed] [Google Scholar]
- 8.Afonnikov DA, Kolchanov NA. CRASP: a program for analysis of coordinated substitutions in multiple alignments of protein sequences. Nucleic Acids Res. 2004;32:W64–W68. doi: 10.1093/nar/gkh451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulka B, desJardins M, Freeland SJ. An interactive visualization tool to explore the biophysical properties of amino acids and their contribution to substitution matrices. BMC Bioinformatics. 2006;7:329. doi: 10.1186/1471-2105-7-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pokarowski P, Kloczkowski A, Jernigan RL, Kothari NS, Pokarowska M, Kolinski A. Inferring ideal amino acid interaction forms from statistical protein contact potentials. Proteins. 2005;59:49–57. doi: 10.1002/prot.20380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyazawa S, Jernigan RJ. Self-consistent estimation of inter-residue protein contact energies based on an equilibrium mixture approximation of residues. Proteins. 1999;34:49–68. doi: 10.1002/(sici)1097-0134(19990101)34:1<49::aid-prot5>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 12.Pokarowski P, Kloczkowski A, Nowakowski S, Pokarowska M, Jernigan RL, Kolinski A. Ideal amino acid exchange forms for approximating substitution matrices. Proteins. 2007;69:379–393. doi: 10.1002/prot.21509. [DOI] [PubMed] [Google Scholar]
- 13.Seto Y, Ihara S, Kohtsuki S, Ooi T, Sakakibara S. Peptide and protein databanks in Japan. In: Lesk AM, editor. Computational Molecular Biology. Oxford: Oxford University Press; 1988. pp. 27–37. [Google Scholar]
- 14.Fujibuchi W, Goto S, Migimatsu H, Uchiyama I, Ogiwara A, Akiyama Y, Kanehisa M. DBGET/LinkDB: an integrated database retrieval system. Pacific Symp. Biocomput. 1998, 1998:683–694. [PubMed] [Google Scholar]
- 15.Kanehisa M. Linking databases and organisms: GenomeNet resources in Japan. Trends Biochem. Sci. 1997;22:442–444. doi: 10.1016/s0968-0004(97)01130-4. [DOI] [PubMed] [Google Scholar]
- 16.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]