Abstract
New methods and tools are needed to exploit the unprecedented source of information made available by the completed and ongoing whole genome sequencing projects. The Narcisse database is dedicated to the study of genome conservation, from sequence similarities to conserved chromosomal segments or conserved syntenies, for a large number of animals, plants and bacterial completely sequenced genomes. The query interface, a comparative genome browser, enables to navigate between genome dotplots, comparative maps and sequence alignments. The Narcisse database can be accessed at http://narcisse.toulouse.inra.fr.
INTRODUCTION
The advent of complete genome sequences raised the needs for genome browsers and visualization tools. The international scientific community has encouraged a collaborative effort of the bioinformatics community to develop such software. This effort was put into concrete form with the well-known browsers at NCBI (1), UCSC (2) and Ensembl (3). These browsers were first developed in order to enable the publication of the entirely sequenced human genome and its annotation. They were subsequently extended to all completely sequenced animal genomes and also more recently to other kingdoms (1). In addition to these general purpose genome browsers, some software have been devised specifically in a comparative genomics perspective: SynBrowse (4), SynView (5) or SYBIL (http://sybil.sourceforge.net) for the representation of conserved syntenies and VISTA (6) for genomic sequence alignments. The first browsers are restricted to the representation of syntenies. In contrast, the VISTA software is dedicated to the representation of sequence conservation but lacks a comprehensive representation of chromosomal rearrangements between the genomes.
The Narcisse database performs the comparative analysis of completely sequenced genomes with a special emphasis on a combined analysis of sequence similarities and structural rearrangements. The Narcisse database and web-based front-end software differ from the existing and previously mentioned software in both its particular treatment of the segment conservation concept, the meaning of which varies with the level of map resolution and in the possibility to navigate and explore the ensuing different levels of genome conservation, from genome comparisons to comparative maps and sequence conservation. The Narcisse database provides an integrated approach for comparing completely sequenced genomes of the animal, plant, fungi and bacterial kingdoms. The mirror-like behaviour of small conserved elements within a larger conserved segment, central to our multilevel representation genome conservation but also to all representations in comparative genomics, suggested the mirror metaphor for the database name.
Comparative mapping in the whole genome sequence era
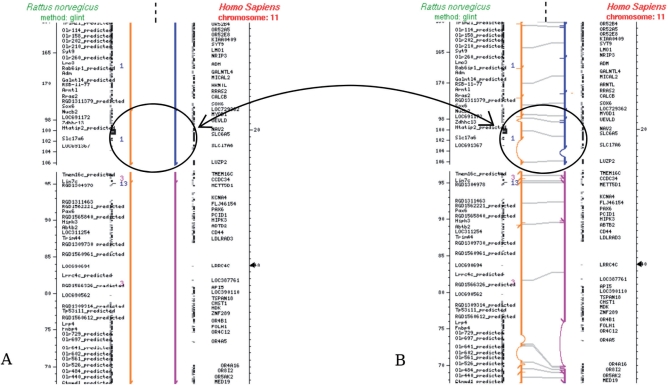
Following (7), we propose to take advantage of the bottom-up approach for conserved segment identification to construct different levels of genome conservation. Starting with local sequence alignments, our algorithm, described in Supplementary Data S1, iteratively constructs conserved segments by chaining conserved segments of a previously defined conservation level according to the respective organization of these elements in both genomes. This introduces a concept of conservation that is dependent on the chromosomal context. At the highest level, the chromosome level, the conserved segments represent chromosomal regions sharing a large number of genes without implying a strict conservation of gene order while at the lowest level, the sequence level, a conserved segment is represented by a sequence similarity attested by a high-scoring local sequence alignment. Intermediate levels are obtained by chaining elements of a lower level with a tolerance to out-of-place segments. An example of two consecutive conservation levels is given in Figure 1.
Figure 1.
Conserved segments. Comparative map between a region of human chromosome 11 and the corresponding conserved segments with rat chromosomes 1, 3 and 13. (A) Representation of 7 conserved segments along human chromosome 11 (right) colour coded according the corresponding rat chromosomes in the following order: 1, 1, 3, 13, 3, 1 and 1. The corresponding rat chromosome regions are represented opposite facing on the left. (B) Decomposition of these segments into chains of segments from a lower level of conservation. The segment circled in A decomposes for example into the 4 segments in B. Note in (B) the variable size of the gaps between conserved segments pinpointing the different evolutionary story that the two lineages have undergone with a large segmental deletion occurring in one of the lineages (or an insertion in the other).
Data sources and processing
For each species, Narcisse uses the complete genome sequence and its associated annotation provided by the public sequence database repositories as raw material. For animal genomes, Narcisse uses the Ensembl API programs (3) to download the latest annotation and complete chromosome sequences. For bacterial and fungus genomes, the GenBank files containing both annotations and sequences are downloaded from the NCBI genome sequence repository (ftp://ftp.ncbi.nih.gov/genomes). The sequences and annotations of plant genomes are directly downloaded from the home pages of each genome project.
Whole genome comparisons
Sequence comparison
We perform whole genome comparisons both at the protein level, a blastp (8) comparison of annotated gene sets, and at the nucleic level. For the latter, we have developed a program, named ‘glint’, based on the construction of an index for each genome (Faraut,T. and Courcelle,E., manuscript in preparation). The output of both comparisons takes the form of a set of local similarities that are transformed, independently, into one-to-one correspondence orthologous landmarks based on a reciprocal best-hit criterion. These orthologous landmarks are subsequently chained in order to provide the conserved segments that make the different levels of chromosome conservation.
Constructing conserved segments
In order to reconstruct conserved chromosomal segments, we proceed in a multiple pass algorithm described in detail in Supplementary Data S1. Briefly, starting with local sequence similarities, conserved segments are chained according to an adaptation of the Longest Increasing Subsequence (LIS) algorithm transformed into a general purpose dynamic programming algorithm that identifies high-scoring chain of segments occurring in the same order in both species. The levels of conservation are arbitrarily fixed from 0 to 5 for animal and fungal genomes and from 0 to 2 for the other kingdoms. The conservation level 0 corresponds to unchained local sequence alignments for the nucleic comparison or reciprocal best hits for the protein comparison.
The Narcisse comparative genome browser
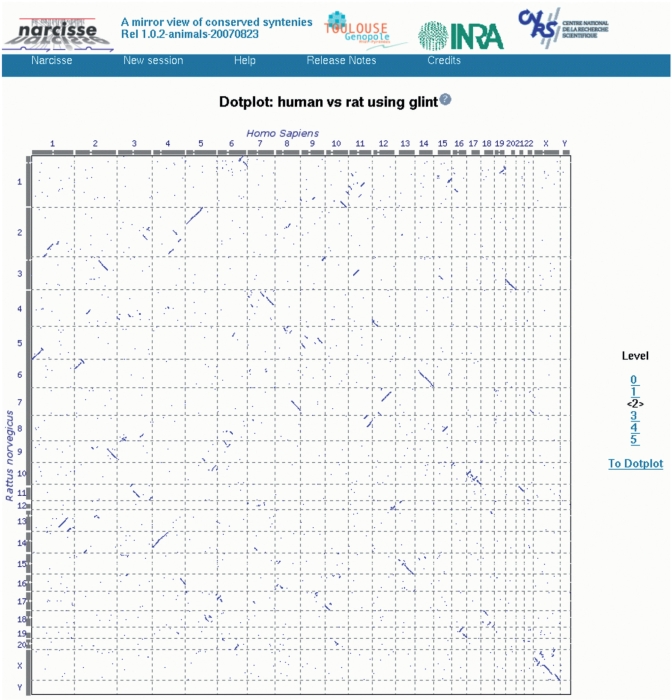
To illustrate the Narcisse functionalities for the exploration of genome conservation we will briefly sketch how, starting from a dotplot representation of the comparison of two genomes, the user can browse the different levels of genome conservation. Starting with the dotplot presented in Figure 2, a chromosome of one of the two organisms can be selected as a reference for the chromosome comparison main view.
Figure 2.
Interactive dotplot. From the right panel, the conservation or chaining level can be selected, from level 0 containing all elementary reciprocal best hits to level 5 for macro syntenies only. A click on the dotplot zooms in on the dotplot of the corresponding pair of chromosomes. Chromosomes IDs and chromosome arm pictures on XY axes are linked to the detailed views of the conservations using the chromosome on the selected axis as the reference sequence.
The chromosome comparison main view
The Narcisse comparative chromosome representation is based on a reference organism, displayed centrally, together with its associated annotation on which the identified conserved segments are anchored (either right or left, Figures 1, 3A and 4). Figure 1A shows a region of human chromosome 11 on which at least 7 segments have been identified as conserved with the rat genome. On each conserved segment, annotation is displayed according to the chromosome coordinate of this segment. As a result of the chaining algorithm, each conserved segment is made of segments of a lower level as exemplified by Figure 1B. The comparative representation of conserved segments can be adapted to the resolution level of the map under consideration; large conserved chromosomal segments are preferred at a cytogenetic scale while the representation of local sequence conservation is more informative when studying the vicinity of a set of genes (Figure 3B).
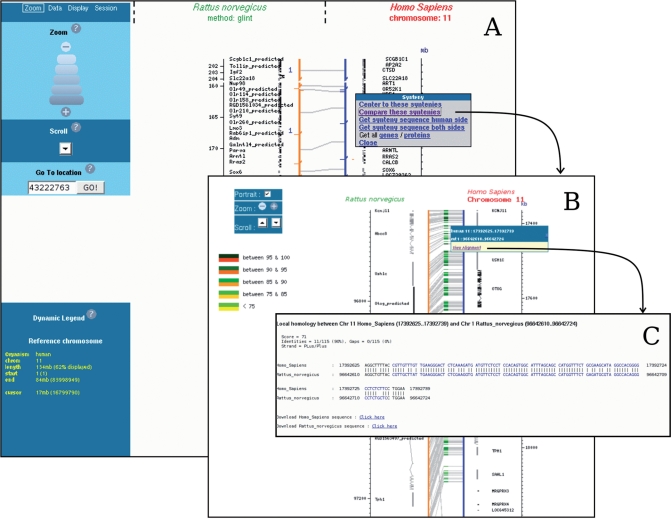
Figure 3.
Narcisse comparison layers; from conserved segments to sequence similarities. The top level layer (A) displays the reference sequence (bold, red labelled) and the corresponding conserved segments on the target genome (e.g. blue bars represent conserved segments with regions of rat chromosome 1). When a conserved segment is selected, a more sensitive comparison can be performed. The output of this online computation is presented in (B). This view presents dna/dna similarities where any local pairwise alignment can be visualized (C). This low level view shows base pair conservations where blue coloured nucleotides encode for protein coding regions.
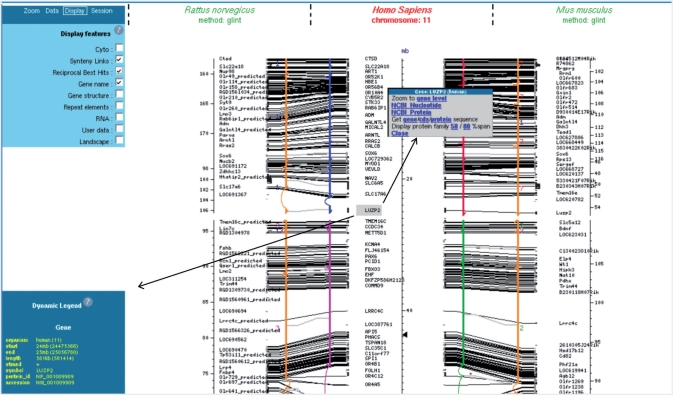
Figure 4.
Customisable layout and data access. Up to three genomes can be compared in the same view (one reference and two targets). The information content of the page is customizable by using the Display tab of the control panel. At any time the user can get the details of a gene element in the dynamic legend or retrieve corresponding sequences (gene/cds or proteins) using the contextual popup menu.
From chromosome conservation to sequence similarities
The chromosome comparison main view enables to explore in a continuous manner the Narcisse data from the whole genome level to the base or amino acid level (Figure 3A, B and C). Starting with the higher level, the genome comparisons are displayed at the chromosome level where not all genes or links are represented, but which provides the user with a synthetic view of the conservation (Figure 3A). Focusing on smaller regions is then possible using one of the available zoom or positioning tools. In order to study in greater detail a particular conserved segment, a click on this segment enables to compute dynamically an alignment between the two segments involved in the conservation (Figure 3B and C).
Managing the display
A sophisticated zoom tool is available through the control panel. It can be used to zoom in and out or scroll through the reference genome chromosome. Some predefined zoom levels are also defined providing a direct access to biological meaningful views. Many display parameters can be set from the control panel (Display tab; see Figure 4, upper left): cytology data (query chromosome only), synteny links, reciprocal best hits and other features may be displayed or hidden. Using the Data tab of the control panel, it is also possible to add or remove a third track (Figure 4).
User session
An important functionality of Narcisse is the possibility to upload personal data. The data will be automatically integrated to the graphics and adapted to each display mode. The file format, very simple, is described on the help page. The session tab in the control panel allows the user to manage his session, which means that some bookmarks can be defined, and that the session directory can be saved and downloaded.
Conclusion and future perspectives
The Narcisse database is an attempt to provide a simple and intuitive access to the comparison of genomes of the animal, plant and bacterial kingdoms. One of the important aspects of genome evolution presently not addressed in Narcisse is segment duplication. The next methodological development will therefore be to enable the identification of paralogous conserved segments within as well as between species and their relationship with gene families. Future developments should also address the possibility to perform and visualize multiple genome alignments.
Periodic updates and availability
The Narcisse database groups animals, plants, fungi and bacteria in four separate instances. Supplementary Data S2 gives a list of the genomes available for each instance. The update process is highly automated, so that the data are kept up to date on a 6-month periodicity basis. The Narcisse public web site is available at the address: http://narcisse.toulouse.inra.fr.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the Génopole Toulouse Midi-Pyrénées and the CALMIP Project for providing us with the necessary computing resources. We also thank Odile Roussot and two anonymous referees for valuable suggestions for improving the manuscript. Finally, we would like to thank Sandrine Dalmar for graphical design assistance. The Narcisse project was supported by the Agence Nationale de la Recherche (GPLA06026G ANR Genoplante and ArcAnge ANR Genanimal project) and by the Génopôle Toulouse Midi-Pyrénées. Funding to pay the Open Access publication charges for this article was provided by ArcAnge ANR Genanimal project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan X, Stein L, Brendel V. Synbrowse: a synteny browser for comparative sequence analysis. Bioinformatics. 2005;21:3461–3468. doi: 10.1093/bioinformatics/bti555. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, Su Y, Mackey AJ, Kraemer ET, Kissinger JC. Synview: a gbrowse-compatible approach to visualizing comparative genome data. Bioinformatics. 2006;22:2308–2309. doi: 10.1093/bioinformatics/btl389. [DOI] [PubMed] [Google Scholar]
- 6.Dubchak I, Ryaboy DV. Vista family of computational tools for comparative analysis of DNA sequences and whole genomes. Methods Mol. Biol. 2006;338:69–89. doi: 10.1385/1-59745-097-9:69. [DOI] [PubMed] [Google Scholar]
- 7.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.