Abstract
The integrated microbial genomes (IMG) system is a data management, analysis and annotation platform for all publicly available genomes. IMG contains both draft and complete JGI microbial genomes integrated with all other publicly available genomes from all three domains of life, together with a large number of plasmids and viruses. IMG provides tools and viewers for analyzing and annotating genomes, genes and functions, individually or in a comparative context. Since its first release in 2005, IMG's data content and analytical capabilities have been constantly expanded through quarterly releases. IMG is provided by the DOE-Joint Genome Institute (JGI) and is available from http://img.jgi.doe.gov.
INTRODUCTION
With ∼20% of the reported genome projects worldwide, DOE-JGI is one of the main production centers of genome sequence data (1). IMG serves as a community resource for comparative analysis and annotation of all publicly available genomes from all three domains of life, in a uniquely integrated context.
Starting with version 2.0 released in December 2006, IMG has employed NCBI's RefSeq (2) as its main source of publicly available genomes. Through regular updates, IMG's data content has grown from a total of 296 genomes in its first version released in March 2005, to a total of 2 878 genomes in the version released in September 2007. New archaeal and bacterial genomes are added to IMG on a quarterly basis: IMG 2.3 (September 2007) has 729 bacterial and 46 archaeal genomes. An increasing number of eukaryotic genomes, viruses (including phages) and plasmids have been also added to IMG in order to increase its genomic context for comparative analysis: IMG 2.3 has 50 eukaryotic genomes, 1661 viruses and 402 plasmids that did not come from a specific microbial genome sequencing project.
IMG's analytical tools have been gradually generalized and enhanced in terms of their usability, analysis flow and performance. These tools allow users to focus on a subset of genes, genomes and functions of interest, and conduct analysis using summary tables, graphical viewers and various methods for comparing genes, pathways and functions across genomes.
DATA CONTENT AND CURATION
Genomes are identified in IMG using an internally generated unique object identifier (OID). In addition, individual genomes are associated with the NCBI Genomes Project Identifier (PID) and taxonomic lineage via NCBI's Taxonomy (domain, phylum, class, order, family, genus, species and strain). For every genome, IMG incorporates its primary genome sequence information recorded in RefSeq including its organization into chromosomal replicons (for finished genomes) and scaffolds and/or contigs (for draft genomes), cross-referenced with their RefSeq accession identifiers, together with computationally predicted protein-coding sequences (CDSs) and some RNA-coding genes. IMG employs RefSeq's gene identifiers to link to other NCBI resources, such as Entrez Gene (3), and in order to establish gene-based correlations with other microbial genome systems, such as Microbes Online (4).
Functional annotation of genes in IMG consists of: (i) protein product names, (ii) protein family and domain characterization, (iii) IMG term assignment and (iv) MyIMG protein annotation. Protein product names are available from RefSeq and typically consist of the function prediction provided by sequence genome centers. Protein family and domain characterization involve associating genes with various functional roles as defined in different controlled vocabularies, such as Enzyme Nomenclature (5), COG clusters (6), Pfam (7), TIGRfam (8), InterPro (9), Kegg Ortholog (KO) terms (10) and Gene Ontology (GO) terms (11). Genes are associated with COGs and Pfams using RPS-BLAST (Reverse Position-Specific BLAST) computation against NCBI's Conserved Domain Database (CDD) (12). EC numbers are computed using RPS-BLAST against the PRIAM database (the following cutoffs are used: max. E-value: 1E−10; min. percent identity along alignment: 45% and min. alignment fraction over PSSM consensus sequence: 70%) (13), as a complement to the (often sparse) native EC numbers collected via RefSeq. UniProt (14) is used to associate genes with additional annotations, such as InterPro, TIGRfam and GO terms, while KEGG is used to establish KO term associations. RNA gene models are synchronized with Rfam (15). Functional roles are further defined by their association with functional classifications including COG functional categories (6), TIGR role categories (8) and the KEGG pathway collection (10).
In order to address problems with the inconsistencies of the protein product names as well as with the current functional classifications (16), genes are further annotated in IMG using a native collection of generic (protein cluster-independent) functional roles called ‘IMG terms’ that are further defined by their association with generic (organism-independent) functional hierarchies, called ‘IMG pathways’. IMG terms and pathways are currently specified by domain experts at DOE-JGI as part of the process of annotating specific genomes of interest and are subsequently propagated throughout the system. Users can add their own protein annotations that are captured under their user name as MyIMG annotations, as described below.
IMG Terms form a hierarchy, whereby the leaves of this hierarchy consist of functional roles for gene products (protein product descriptions) assigned to individual genes. These lower-level IMG Terms of type ‘Gene Product’ can be directly associated with reactions, whereby they function as either ‘Catalysts’ or ‘Reactants’. Alternatively, they can be assigned recursively as ‘children’ of IMG Terms of type ‘Protein Complex’, thus indicating that they constitute subunits of a multi-subunit protein complex. A detailed discussion of the rationale for IMG terms and pathways and their specification is available at http://img.jgi.doe.gov/pub/doc/imgterms.html, as part of IMG's online documentation. Note that, despite somewhat similar nomenclature, IMG Terms are not equivalent to GO terms (11). A mapping of IMG terms to GO terms is currently developed by the GO consortium in collaboration with DOE-JGI scientists.
Sequence similarities for identifying candidate homologs are computed using NCBI BLASTp with 1E−2 E-value cutoff, and low complexity soft masking (-F ‘m S’) turned on. IMG provides support for filtering candidate homolog lists by percent identity, bit score and more stringent E-values, as well as with a variety of metadata such as phenotype, habitat, etc. In addition, CRISPR repeats (17), signal peptides using SignalP (18) and transmembrane helices using TMHMM (19) are computed, and potentially missing data from the original RefSeq data files (such as various RNAs) are added.
DATA ANALYSIS
Genome data analysis in IMG consists of operations involving genomes, genes and functions that can first be selected and then explored individually. Genomes can be also ‘compared’ in terms of various statistics, gene content, function capabilities and sequence conservation.
Data selection tools
In order to perform comparative analysis in IMG, genomes, genes or functions are first selected using browsers or search tools. Browsers are provided for selecting genomes and functions, organized as alphabetical lists or hierarchically (e.g. based on phylogenetic tree for genomes). Keyword search tools allow identifying genomes, genes and functions of interest using a variety of keyword filters. Genomes can be also selected using a search tool that allows specifying conditions involving phenotype, habitat, disease and relevance metadata fields, while genes can be also selected using BLAST search tools against various datasets. The genomes that result from search operations are displayed as a list from which they can be selected and saved for further analysis. In a similar manner, the genes and functions that result from search operations are displayed as lists from which genes and functions can be selected for inclusion into the ‘Gene Cart’ and ‘Function Cart’, respectively.
Individual genomes can be explored using the ‘Organism Details’ page that includes information on the organism together with various genome statistics of interest, such as the number of genes that are associated with KEGG, COG, Pfam, InterPro or enzyme information. For each genome, one can also examine the associated list of scaffolds and contigs using the ‘Chromosome Viewer’, or can generate circular chromosomal maps on which a variety of data can be projected.
Individual genes can be analyzed using the ‘Gene Details’ page that includes Gene Information, Protein Information and Pathway Information tables, evidence for functional prediction, COG, Pfam and pre-computed homologs. A gene can be examined in the context of its location on the chromosome using the ‘Chromosome Viewer’.
Individual functional groups, such as COG categories, can be further explored using summary pages, such as the ‘COG Category Details’ page that lists the COGs of a given category and the number of organisms that have genes belonging to each COG, where the’organism counts’ are linked to a list of organisms and their associated ‘gene counts’.
Comparative analysis tools
Comparative analysis of genomes is provided in IMG through a number of tools that allow genomes to be compared in terms of various statistics, gene content, function capabilities and sequence conservation.
‘Genome Statistics’ provides statistics across the genomes that have been previously selected and saved as discussed above. The display can be configured by including a variety of genome attributes, such as GC content, number of protein coding genes and various functional annotations.
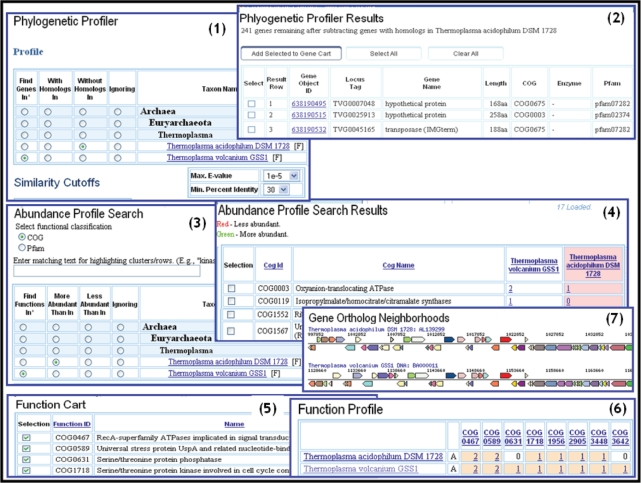
Genomes can be compared in terms of gene content using the ‘Phylogenetic Profiler’ tool that allows to define a ‘profile’ for the genes of the query genome, say the archaeal genome Thermoplasma volcanium GSS1 (T. volcanium) in terms of presence or absence of homologs in any other genomes. In the example shown in pane (1) of Figure 1, the tool is used to find T. volcanium genes that have no homologs in Thermoplasma acidophilum DSM 1728 (T. acidophilum). Similarity cutoffs can be used to fine-tune the selection. The list of genes with the specified profile are then provided as a selectable list as shown in pane (2) of Figure 1. The ‘Phylogenetic Profiler’ tool can be used, e.g. for finding ‘unique’ genes in the query genome with respect to other genomes of interest. In the example shown in Figure 1, 241 genes are found to be unique in T. volcanium with respect to T. acidophilum.
Figure 1.
Profile search and functional profile tools. The ‘Phylogenetic Profiler’ tool is used to find genes in the archaeal genome Thermoplasma volcanium GSS1 (T. volcanium) that have no homologs in a phylogenetically closely related genome, Thermoplasma acidophilum DSM 1728 (T. acidophilum) (1). Similarity cutoffs are used to fine-tune the selection. The list of T. volcanium genes that satisfy the specified condition are provided as a list (2) from which genes can be selected and added to a Gene Cart. The ‘Abundance Profile Search’ tool is used to find COGs that are more abundant in T. volcanium than in T. acidophilum (3). The COGs that satisfy the specified condition are provided as a list (4) from which COGs can be selected and added to the Function Cart. For COGs of the ‘Signal transduction mechanisms’ in the Function Cart, the ‘Function Profile’ tool allows computing a profile across the T. volcanium and T. acidophilum genomes (5). The functional profile result is displayed as a matrix with each cell containing a link to the associated list of genes and displaying the count (abundance) of genes in this list. Colors are used to represent visually gene abundance, whereby white, bisque and yellow represent gene counts of 0, 1–4 and over 4, respectively (6). The T. volcanium and T. acidophilum genes associated with COG0467 are further examined with the ‘Gene Ortholog Neighborhoods’ tool (7): the compared genes are colored red, and their respective neighborhoods appear above and below a single line showing the genes reading in one direction on top and those reading in the opposite direction on the bottom; genes associated with the same color indicate association with the same COG group.
Genomes can be compared in terms of functional capabilities using the ‘Abundance Profile Search’ tool that allows defining a ‘profile’ for functions (COGs, Pfams) in a query genome in terms of their abundance compared to other related genomes.
In the example shown in pane (3) of Figure 1, this tool is used to find COGs that are more abundant in T. volcanium than in T. acidophilum. Some of the COG representatives found in T. volcanium (e.g. COG 1552) have no match in T. acidophilum, which may be of evolutionary significance or explained by the fact that the genes were missed by the original annotation. For each genome, a link to the list of genes associated with individual functions allows examining gene details.
The functional capabilities of genomes can be also compared using a number of additional functional profile tools. First, functions of interest, such as protein families, enzymes and IMG terms, are included into the ‘Function Cart’, as illustrated in pane (5) of Figure 1. For these functions a profile across genomes can be computed, with the results displayed in a tabular format, as illustrated in pane (6) of Figure 1, with each column displaying the profile of a specific function across the genomes. The example in pane (6) of Figure 1 shows the profiles of several COGs of the ‘Signal transduction mechanisms’ COG category across the T. volcanium and T. acidophilum genomes. Each cell in the profile result table displays the count (abundance) of genes in an organism and contains a link to the associated list of genes. Colors are used to represent visually gene abundance, whereby white, bisque and yellow represent gene counts of 0, 1–4 and over 4, respectively. The genes associated with a specific function can be saved using the ‘Gene Cart’ and further examined using various tools, such as gene neighborhood analysis and multiple sequence alignment tools. For example, the ‘Gene Ortholog Neighborhoods’ tool can be used to examine genes of T. acidophilum associated with a specific function (e.g. COG0467) together with its T. volcanium ortholog and their respective chromosome neighborhoods, as shown in pane (7) of Figure 1.
Another functional profile tool, the ‘Abundance Profile Viewer’, provides an overview of the relative abundance of protein families (COGs and Pfams) and functional families (Enzymes) across selected genomes, with abundance of protein/functional families displayed as a heat map. Note that the ‘Function Cart’ in IMG provides users with the opportunity to define their own ‘pathways’ and functional categories, assembled from individual COGs, Pfams or Enzymes. Such user-defined ‘pathways’ can be then employed in analysis of genomes and/or physiological traits that are poorly characterized by the traditional pathway databases, such as KEGG.
Comparative analysis of genes includes gene neighborhood analysis, phylogenetic occurrence profile analysis and multiple sequence alignment, which can be applied to genes collected into the ‘Gene Cart’.
Finally, DNA conservation can be explored for closely related organisms in IMG using the VISTA comparative genome analysis tools (20). Selecting an organism from a predefined list invokes the VISTA browser that can be then used for examining conservation.
User annotations
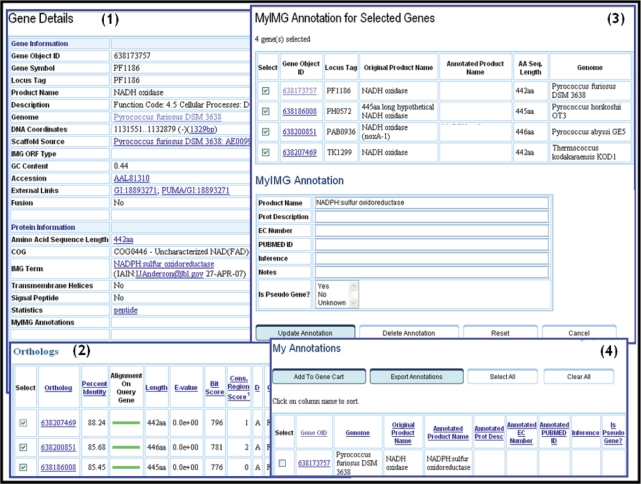
IMG users can enter their own functional annotations using ‘MyIMG’ tools, as illustrated in Figure 2. In this example, a gene of Pyrococcus furiosus is associated with product name NADH oxidase, as shown in pane (1) of Figure 2, and as recorded in GenBank and RefSeq. Based on a recent study (21), it has been determined that the function for this gene is NADPH:sulfur oxidoreductase, and an expert review of the best homologs of this gene indicated that this product name also may be confidently applied to the top three homologs, as shown in pane (2) of Figure 2. The product name and several other gene attributes, such as the associated EC number, can be changed using ‘MyIMG Annotation’ tool, as illustrated in pane (3) of Figure 2. User annotations are stored in IMG and can be reviewed at any time using the same tool. This tool also allows importing user annotations from user files (e.g. from excel files) into IMG or exporting user annotations in IMG to user files.
Figure 2.
MyIMG user annotations. A gene of P. furiosus (with the object identifier ‘638173757’) is associated with product name NADH oxidase, as displayed by its ‘Gene Details’ page (1), and as recorded in GenBank and RefSeq, which can be viewed by following the appropriate External Links. For gene ‘638173757’, one can display its list of orthologs, which can be accessed in the Homologs section of its ‘Gene Details page (2). The top three orthologs of gene ‘638173757’ can be selected and saved together with the gene into the Gene Cart, where the product name is changed to NADPH:sulfur oxidoreductas with the ‘MyIMG Annotation’ tool (3). Other annotations (e.g. EC number) can be also modified. User annotations can be subsequently reviewed, imported into IMG from a user file, or exported to a user file using ‘My Annotations’ page (4).
FUTURE PLANS
IMG continues to be extended in terms of data content through quarterly updates, whereby it aims at continuously increasing the number of genomes integrated in the system from public and local resources, following the principle that the value of genome analysis increases with the number of genomes available as a context for comparative analysis.
Future versions of IMG will focus on further improving the quality of gene models and functional annotations. We plan to expand the native IMG term controlled vocabulary and IMG pathway classification, jointly with annotation of IMG genomes using these terms and pathways. We also plan to provide extensive corroboration of annotations from other public microbial genome data resources, by including into IMG annotations based on TIGR Genome Properties (8) and MetaCyc (22). New data types such as results from microarray and proteomic experiments, as well as information on transcriptional regulatory binding sites will be also included into IMG.
IMG's analytical tools will continue to be extended in order to address two main challenges. First, as IMG's content expands, improved viewers will be developed in order to facilitate the exploration of a rapidly increasing number of genomes, genes and annotations. Additional tools and viewers for exploring the power of gene context (i.e. fusions and gene neighborhood) are also under current development. Since the comparative analysis context provided by IMG helps detect gene model and annotation errors, user annotation tools will be further extended based on requirements and feedback from the user community.
ACKNOWLEDGEMENTS
We thank, Philip Hugenholtz, Anu Padki, Kristen Taylor, Alla Lapidus and Paul Richardson for their contribution to the development and maintenance of IMG. The work of JGI's production, cloning, sequencing, assembly, finishing and annotation teams is an essential prerequisite for IMG. Chris Oehmen of the Computational Biology and Bioinformatics group at the Pacific Northwest National Laboratory provided invaluable help in carrying out the large-scale gene similarity computations for IMG 2.0. Eddy Rubin and James Bristow provided, support, advice and encouragement throughout this project. The work presented in this article was performed under the auspices of the US Department of Energy's; Office of Science, Biological and Environmental Research Program and by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396. Funding to pay the Open Access publication charges for this article was provided by Department of Energy Joint Genome Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Liolios K, Tavernarakis N, Hugenholtz P, Kyrpides N. The genomes online database (GOLD) v.2: a monitor of genome projects worldwide. Nucleic Acids Res. 2006;34:D332–D334. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts, and proteins. Nucleic Acid Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maglott DR, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acid Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alm EJ, Huang KH, Price MN, Koche RP, Keller K, Dubchak IL, Arkin AP. The Microbes Online web site for comparative genomics. Genome Res. 2005;15:1015–1022. doi: 10.1101/gr.3844805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatusov RL, Koonin EV, Lipman DJA. Genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 7.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, Richter AR, White O. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bradley P, Bork P, Bucher P, et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gene Ontology Consortium. The Gene Ontology Database and Informatics Resource. Nucleic Acids Res. 2004;32:258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claudel-Renard C, Chevalet C, Faraut T, Daniel Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31:6633–6639. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanova NN, Anderson I, Lykidis A, Mavrommatis K, Mikhailova N, Chen IA, Szeto E, Palaniappan K, Markowitz V, et al. Technical report 62292. Lawrence Berkeley National Laboratory; Metabolic reconstruction of microbial genomes and microbial community metagenomes. [Google Scholar]
- 17.Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P. CRISPR Recognition Tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 19.Moller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 20.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schut GJ, Bridger SL, Adams MW. Insights into the metabolism of elemental sulfur by the hyperthermophilic archaeon Pyrococcus furiosus: characterization of a coenzyme a-dependent NAD(P)H Sulfur Oxidoreductase. J. Bacteriol. 2007;189:4431–4441. doi: 10.1128/JB.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caspi R, Foerster H, Fulcher CA, Hopkinson R, et al. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2006;34:D511–D516. doi: 10.1093/nar/gkj128. [DOI] [PMC free article] [PubMed] [Google Scholar]