Abstract
MicroRNAs have been found in various organisms and play essential roles in gene expression regulation of many critical cellular processes. Large-scale computational prediction of miRNAs has been conducted for many organisms using known genomic sequences; however, there has been no such effort for the thousands of known viral genomes. Some viruses utilize existing host cellular pathways for their own benefit. Furthermore, viruses are capable of encoding miRNAs and using them to repress host genes. Thus, identifying potential miRNAs in all viral genomes would be valuable to virologists who study virus–host interactions. Based on our previously reported hairpin secondary structure and feature selection filters, we have examined the 2266 available viral genome sequences for putative miRNA hairpins and identified 33 691 hairpin candidates in 1491 genomes. Evaluation of the system performance indicated that our discovery pipeline exhibited 84.4% sensitivity. We established an interface for users to query the predicted viral miRNA hairpins based on taxonomic classification, and a host target gene prediction service based on the RNAhybrid program and the 3′-UTR gene sequences of human, mouse, rat, zebrafish, rice and Arabidopsis. The viral miRNA prediction database (Vir-Mir) can be accessed via http://alk.ibms.sinica.edu.tw.
INTRODUCTION
MicroRNAs (miRNAs) are endogenous non-protein-coding RNAs that are ∼22-nt long. They negatively regulate gene expression by complementary binding to the 3′-UTR regions of target genes (1). Since the first discovery of miRNA in Caenorhabditis elegans, thousands of miRNAs have been computationally and/or experimentally identified in many organisms, including mammals, invertebrates, insects and plants (2). It has been shown that miRNAs can function in various physiological pathways. Plant miRNAs can regulate development in embryos, leaves and floral meristems (3,4). Mammalian miRNAs participate not only in developmental regulation (5) but also in pathogenesis if they are dysfunctional (6). Even with limited information about miRNA gene structures and their target genes’ recognition selections, several genome-wide bioinformatics approaches have been recently employed to identify these important molecules (7–9). Establishing such a bioinformatic resource for miRNA will be beneficial for daily experimental research in the biological laboratories.
Similar to many eukaryotic organisms, viruses also encode miRNAs (10,11). To date, at least 82 viral-associated miRNAs have been identified from eight different viruses (2). Because viruses are often parasitic, these viral miRNAs may target important host genes to reduce the host cell defense and to control host cell biogenesis. Human herpes virus 4 (Epstein-Barr virus, EBV) represses several host genes, including those encoding B cell-specific chemokines and cytokines, transcriptional regulators and components of signal transduction pathways, by means of virus-encoded miRNAs (11). Moreover, HIV-1 also has been proposed to enhance viral infection capability by repressing host immune system genes via specific HIV viral miRNAs (10).
MOTIVATION
It would be beneficial for biomedical researchers who study virus–host interactions to identify potential viral miRNAs and their target genes in hosts. Although a new database collecting viral gene targets of host miRNAs has been reported recently (12), but there is no genome-wide miRNA prediction for all completed viral genome sequences. We have modified our previous miRNA discovery pipeline to predict viral miRNA hairpins (13). Here, we present our research effort on candidate hairpin discovery of virus-encoded miRNAs from all virus genomes available from the National Center for Biotechnology Information (NCBI), National Institutes of Health, USA. It would be beneficial to have a user-friendly web interface to query the predicted viral miRNA hairpins based on their familial taxonomic classification. In addition, a host target gene prediction service is established based on the RNAhybrid program and the parsed 3′-UTR gene sequences of human, mouse, rat, zebrafish, rice and Arabidopsis species to search for potential target genes of user's interest.
DATA GENERATION
Predicting hairpins from virus genome scaffold sequences using Srnaloop
During miRNA maturation, the full-length pre-miRNA transcript needs to form a hairpin (stem-loop) structure. The secondary structure is folded via intramolecular base pairing and has been the most significant criterion for computational identification of miRNAs (13–16). The Srnaloop program, developed by Grad et al. (14), has been used to identify putative hairpin secondary structures from viral genomic sequences. All the viral genome sequences were obtained on 4 May 2006 from NCBI, National Institutes of Health, USA. The NCBI viral1.genomic.fna file comprises the genomic sequences of 2266 viruses, and the file size is 41 MB. The optimized parameters for Srnaloop were as reported previously (13), except for the parameters ‘–l 90’ and ‘–t 17’. These parameters, modified based on known viral pre-miRNAs, are specific for identifying hairpins that are up to 90 bases long and have a score of at least 17.
Sequence and structural features filter
To reduce the number of falsely predicted miRNAs based on hairpin structure alone, we have included several additional selection filters, namely GC content, minimum free energy of the core of hairpin structure (core mfe), minimum free energy of the whole hairpin (hairpin mfe) and the ratio of core mfe to hairpin mfe (ch_ratio) as defined in a previous publication (13). To distinguish authentic miRNA candidates, we also employed additional classification criteria specific to known miRNAs. We first investigated the features of known viral miRNAs as well as their precursors and then determined the reference range values of the selected features. The reference range values are listed in Table 1. Therefore, a candidate hairpin with sequence and structural features within the optimized reference range values was considered to be a positive miRNA hairpin.
Table 1.
Distribution range of sequence and structure features from known viral miRNAs
| GC content | core mfe | hairpin mfe | ch ratio | |
|---|---|---|---|---|
| Distribution | ∼34–65 | −37.5 to −14.5 | −22 to −59.2 | ∼51–100 |
| Reference value | ∼38–65 | −37.5 to −19.4 | −50.9 to −28.2 | ∼51–98 |
From the known viral pre-miRNA information, we evaluated the distributions of quantifiable sequence and structure features, namely GC content, core mfe, hairpin mfe and the ratio of core mfe to hairpin mfe. Because extreme values existed, we adopted the reference value to be used in the sequence and structure feature filter.
Application of open reading frame (ORF) feature filter
When predicting miRNAs in organisms other than viruses, a ‘conservation pattern’ is usually used to find evolutionarily conserved miRNA candidates (13–16). However, this feature is not applicable for most viral miRNA prediction here. Cullen found only a few miRNAs located in protein-coding transcripts (17), which is used as a criterion to predict eukaryotic miRNAs (13,14). Therefore, we screened our candidate hairpins to check if they overlap the ORFs belonging to the same virus. The sequences of the ORFs were extracted from viral1.protein.gbff file downloaded from NCBI on 4 May 2006. Because viruses encode functional genes redundantly in a highly compact genomic region, we cannot exclude the possibility that viral miRNAs may overlap other viral protein-coding genes. This concern is more significant for small genome RNA viruses. Therefore, we still considered the hairpins overlapping ORFs to be positive miRNA hairpins, but we marked the NP accession numbers of the overlapped ORF protein in the dataset for reference.
DATABASE STATISTICS
Candidate hairpins predicted by Srnaloop and additional feature filters
We identified roughly 514 874 candidate hairpins from all viral genomic sequences by means of Srnaloop from 2266 viral genomes. They were then divided into 5P and 3P stem arms as putative miRNA candidates as done previously (13), because an miRNA could be located in either the 5' arm or 3' arm of the stem-loop precursors. Following the screen with the sequence and structure feature filter, 33 691 candidates survived. Among them, 5306 candidate hairpins do not overlap any ORF of the correspondent virus, implying they have higher likelihood to be authentic, but not absolutely as described earlier.
All the hairpin information can be accessed from the web interface. We compared the sequences of these putative viral candidate miRNAs with the sequences of known miRNAs in miRBase (release 9.0). The positive match criterion required at least an 18-nt matched length with >90% identity. With this high stringency of sequence similarity, only seven candidates with unique IDs of 821661, 1341202, 1343051, 1408482, 1430283, 3325175 and 3663618 showed positive matches. For example, candidate 821661, identified from Cercopithecine monkey herpes virus 15, is similar to ebv-miR-BART19; candidates 1341202, 1343051 and 3325175, identified from chimpanzee cytomegalovirus, are similar to hcmv-miR-UL112, hcmv-miR-UL148D and hcmv-miR-UL112, respectively. It is possible that more matches could be found if the stringency of sequence similarity was lowered.
Discovery pipeline performance
To assess the efficacy of our pipeline system, we performed both sensitivity and specificity tests. Because it is much easier to identify hairpin structures in sequences of known hairpins than from entire genomic sequences, we calculated the number of predicted candidate hairpins after each filter procedure in our pipeline to determine the recovery rate of reported miRNAs. Originally, the test dataset consisted of 64 known pre-miRNAs belonging to five viruses from miRBase. As shown in Table 2, 63 known miRNAs remained after the Srnaloop step, 54 after the sequence and structure features step, and 50 after the ORF comparison step of the selection. These results imply that the recovery rate is close to 98% after Srnaloop hairpin prediction, 84% after Srnaloop hairpin prediction and sequence and structure feature filter and 78% after additional ORF filter.
Table 2.
System performance test and the number of candidate hairpins
| Known viral miRNAs (species) | Number of miRNAs | Number of miRNAs after Srnaloop filter | Number of miRNAs after sequence and structure filter | Number of miRNAs after ORF filter |
|---|---|---|---|---|
| HCMV | 11 | 11 | 8 | 6 |
| KSHV | 13 | 12 | 8 | 7 |
| RLCV | 16 | 16 | 14 | 14 |
| SV40 | 1 | 1 | 1 | 0 |
| EBV | 23 | 23 | 23 | 23 |
| Total (Sensitivity) | 64 (100%) | 63 (98.4%) | 54 (84.4%) | 50 (78.1%) |
| Total number of predicted miRNA candidate hairpins | 2266 virus genomic sequences | 514 874 | 33 691 | 5306 |
To test the performance of our system, we calculated the number of known viral pre-miRNAs remaining after each filter process. Originally, there were 64 known pre-miRNAs belonging to five viruses. The number of hairpins predicted from 2266 virus genomes is also listed.
For the specificity examination on the pipeline, the negative dataset generation procedure was similar to the one described by Sewer et al. (18). This procedure is based on the fact that the fraction of miRNA-encoding sequences in the genome is very small; therefore, randomly extracted sequences are extremely unlikely to encode miRNAs. We randomly extracted 19 200 (64 × 300) sequence fragments (90 bp in length) from viral genomic sequences. These 19 200 randomly chosen sequences of 1.8 Mb were applied to our discovery pipeline under the same hairpin identification parameters and sequence and structural filter criteria. As a result, we obtained 533 presumed false positives from three independent experiments (175, 176 and 182 predicted candidates, respectively), corresponding to an average of 178 false positives. This would give an estimated 1% false positive prediction in a randomly generated 20 000 sequences. With over 40 000 000 nt from virus genomes, we could estimate ∼4000 false positive predictions (using non-overlapping 90 bp length windows). Therefore, the best specificity we could achieve is close to 88%, and the actual prediction rate would lower depending on the viral genome structures (DNA versus RNA viruses) and gene organizations.
WEB INTERFACE FOR VIR-MIR db
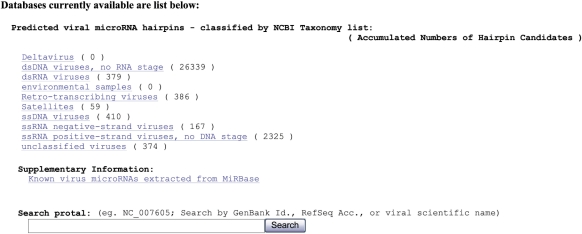
Due to the large number of prediction results, we constructed a user-friendly web interface to present the viral candidate miRNAs. As shown in Figures 1 and 2, the classification and arrangement of the viral candidates are according to the taxonomy table of NCBI, so users can query the viral miRNAs more easily. Users may query the putative miRNAs from a specific virus by using the hierarchical menu or by using the simple search function. A keyword search can be performed, but users are recommended to use it with the GenBank identifier or RefSeq accession number for better search result, e.g. 30844336 or NC_003663 for Cowpox virus.
Figure 1.
Viral miRNA candidate hairpin query web interface. Users may query the putative miRNAs of a specific virus by using the hierarchical menu or by using the search function with GenBank identifier, RefSeq accession number or keyword.
Figure 2.
Expanded view of taxonomy table. The classification and arrangement of viral miRNA candidate hairpins are based on the taxonomy table of NCBI. The numbers in the right parentheses of each classification level indicate the numbers of candidate hairpins found in those levels. By clicking on a link, users can see the expanded table. The numbers in parentheses indicate the number of candidate hairpins found in that particular viral genome.
The numbers in the right parentheses of each classification level indicate the numbers of candidate hairpins found in that level. For example, Atadenovirus has 18 candidate hairpins, which are cumulated from its four sub-classes of viruses, including Bovine adenovirus, Duck adenovirus A, Ovine adenovirus D and Possum adenovirus. We identified 16 candidate hairpins from Duck adenovirus A and no candidate hairpin from Bovine adenovirus D. In some cases, like Bovine adenovirus 4, 5, 8 and Possum adenovirus, we did not record the number of candidate hairpins in their right parentheses, which means there are no genomic sequences available for analysis at these classification levels.
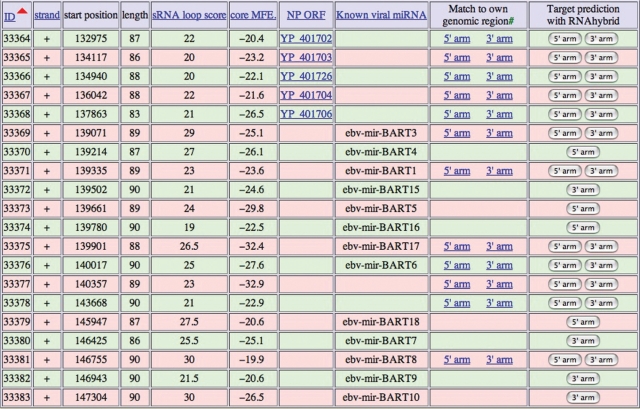
After selecting a specific virus type, as illustrated in Figure 3 for human herpes virus 4, users may further sort the candidate hairpins according to the miRNA genomic location (default), score by Srnaloop, or core mfe by RNAfold. Moreover, for known pre-miRNAs, the sequences and locations of mature miRNAs are shown according to the known miRNA information. For example, both ebv-mir-BHRF1-1 and ebv-mir-BHRF1-3 have only one mature miRNA in their 5′ arms. However, the pre-miRNA ebv-mir-BHRF1-2 has two mature miRNAs in its 5′ arm and 3′ arm. In addition, users may download the entire sequences of predicted candidate hairpins via the link at the bottom of each page. Besides, we also extracted all known viral miRNAs from miRBase into an independent table that also can be retrieved at http://alk.ibms.sinica.edu.tw/cgi-bin/miRNA/known_viral_miRNA.cgi.
Figure 3.
Viral miRNA candidate hairpin web interface. Here, users can view all the predicted viral miRNA hairpin information for a particular virus. They may sort the candidate hairpins according to genomic location (default), score calculated by Srnaloop or core mfe calculated by RNAfold. The YP accession number in the NP reference protein column denotes the protein-coding ORF sequence overlapped by this candidate hairpin. Click the field header to sort on a field. Click it again to reverse the sorted list.
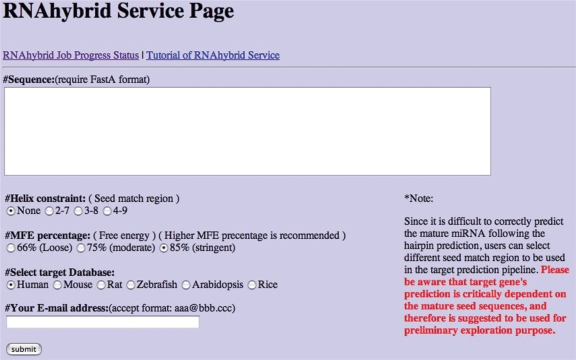
Predicting potential viral miRNA target genes using the RNAhybrid program
It is of researchers’ interest to learn about viral miRNAs and their host target genes. To assist biologists, we provide an integrated interface to identify possible host target genes of the viral miRNA in selected host genomes using the RNAhybrid program (19,20). Presently, as shown in Figure 4, we provided the 3′-UTR region sequences of reference genes from human, mouse, rat, zebrafish, rice and Arabidopsis as the search target dataset. We retrieved 3′ UTRs based on the coding sequence positions acquired individually from the human.rna.gbff file (from NCBI on 19 December 2005), mouse.rna.gbff file (from NCBI on 19 December 2005), rat.rna.gbff file (from NCBI on 19 December 2005), zebrafish.rna.gbff file (from NCBI on 19 June 2006) and plant1 ∼ 5.rna.gbff file (from NCBI on 5 August 2006).
Figure 4.
Target gene search web interface. Presently, we provide the 3′-UTR region sequences of human, mouse, rat, zebrafish, rice and Arabidopsis for target gene prediction. The RNAhybrid program is applied for the target search (19,20). Users can directly submit the predicted viral miRNA hairpin sequences from the viral miRNA web interface or submit their own miRNA sequences for target gene prediction. We have a helix constraint option to let users select different seed match region to be used in the target prediction pipeline. Since it is difficult to correctly predict the mature miRNA from the hairpin structure. It is suggested this target prediction information should be used for preliminary exploration purpose.
Users may scan the 3′-UTR region sequences of reference genes for possible viral miRNA targets. When operating RNAhybrid, the pipeline first calculates the optimal free energy of a putative miRNA when the entire putative miRNA binds to a perfectly complementary target site, then it calculates the minimum free energy of RNA duplex (mfe of the miRNA/mRNA duplex). An alignment for which the RNA duplex mfe is more than 66% of its correspondent optimal free energy is regarded as a positive alignment as described by Krek et al. (21). A higher stringent 85% parameter is recommend. A critical issue on the target prediction is that it dramatically depends on the seed region located in the mature miRNA. However, it is difficult to correctly predict the mature miRNA following the hairpin prediction pipeline. Therefore, we have a helix constraint option to let users select different seed match region to be used in the target prediction pipeline. It is suggested this information should be used for preliminary exploration purpose without the mature miRNA information.
DISCUSSION
The available predicted viral miRNA candidate hairpins will certainly be beneficial to biologists who study interactions between viral miRNAs and host genes. There are many completed viral genome sequences in the database, but it is not a simple task for biologists to execute the entire bioinformatic pipeline to scan for putative miRNA hairpins. We have identified 33691 candidate hairpins from more than 2000 virus genome sequences. Here, we provide an easy-to-use web interface for examining predicted viral miRNA hairpins based on the previously published pipeline. To make this tool more user-friendly, we also included the target gene search function. Many functionally related miRNAs cluster together to facilitate their expression and functional regulation. For example, about half of the miRNAs identified in Drosophila are reported to exhibit this clustering phenomenon; they are initially co-transcribed from one polycistronic transcript and further processed into distinct individual mature miRNAs (1,22,23). This phenomenon has been reported in EBV (11) and also was observed in our present results. As illustrated in Figure 3, two predicted candidate hairpins from this study are clustered in the transcript of BART together with several known EBV pre-miRNAs (11). This phenomenon is also found in the transcript of BHRF1, which contains two newly predicted candidate hairpins and three previously known pre-miRNAs, ebv-mir-BHRF1-1, ebv-mir-BHRF1-2 and ebv-mir-BHRF1-3. This clustering phenomenon indicates that these candidate hairpins may be authentic viral miRNAs.
ACKNOWLEDGEMENT
This research is partial supported by the Academia Sinica research grant support. Funding to pay the Open Access publication charges for this article was provided by the National Science Council, Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 6.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. miRGen: a database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2006:D149–155. doi: 10.1093/nar/gkl904. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu PW, Huang HD, Hsu SD, Lin LZ, Tsou AP, Tseng CP, Stadler PF, Washietl S, Hofacker IL. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–D139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennasser Y, Le SY, Yeung ML, Jeang KT. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology. 2004;1:43. doi: 10.1186/1742-4690-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 12.Hsu PW, Lin LZ, Hsu SD, Hsu JB, Huang HD. ViTa: prediction of host microRNAs targets on viruses. Nucleic Acids Res. 2007;35:D381–D385. doi: 10.1093/nar/gkl1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li SC, Pan CY, Lin WC. Bioinformatic discovery of microRNA precursors from human ESTs and introns. BMC Genomics. 2006;7:164. doi: 10.1186/1471-2164-7-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grad Y, Aach J, Hayes GD, Reinhart BJ, Church GM, Ruvkun G, Kim J. Computational and experimental identification of C. elegans microRNAs. Mol. Cell. 2003;11:1253–1263. doi: 10.1016/s1097-2765(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 15.Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 17.Cullen BR. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Sewer A, Paul N, Landgraf P, Aravin A, Pfeffer S, Brownstein MJ, Tuschl T, van Nimwegen E, Zavolan M. Identification of clustered microRNAs using an ab initio prediction method. BMC Bioinformatics. 2005;6:267. doi: 10.1186/1471-2105-6-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 22.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 23.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]