Abstract
MicroRNA.org (http://www.microrna.org) is a comprehensive resource of microRNA target predictions and expression profiles. Target predictions are based on a development of the miRanda algorithm which incorporates current biological knowledge on target rules and on the use of an up-to-date compendium of mammalian microRNAs. MicroRNA expression profiles are derived from a comprehensive sequencing project of a large set of mammalian tissues and cell lines of normal and disease origin. Using an improved graphical interface, a user can explore (i) the set of genes that are potentially regulated by a particular microRNA, (ii) the implied cooperativity of multiple microRNAs on a particular mRNA and (iii) microRNA expression profiles in various tissues. To facilitate future updates and development, the microRNA.org database structure and software architecture is flexibly designed to incorporate new expression and target discoveries. The web resource provides users with functional information about the growing number of microRNAs and their interaction with target genes in many species and facilitates novel discoveries in microRNA gene regulation.
INTRODUCTION
Mature microRNAs are a class of small, single-stranded RNAs involved primarily in the negative regulation of gene expression. MicroRNA-mediated regulation is a key component in a wide range of biological processes such as stem cell maintenance, developmental timing, metabolism, host–viral interaction, apoptosis, cardiac and skeletal muscle proliferation (1) and neuronal gene expression (2). A number of human diseases including cancer are associated with changes in the copy number or expression of microRNAs and with mutations in the sequence of microRNAs or their target sites. The importance of microRNAs is further underscored by their ubiquitous expression in almost all cell types and their evolutionary conservation in most of metazoan and plant species.
MicroRNAs act as adaptors that employ a silencing complex to target mRNAs by selective base-pairing, primarily in the 3′-UTR region. Target interaction does not require perfect complementarity between microRNA and mRNA sequences, although near-prefect base-pairing in a small region in the 5′end (positions 2–8) of the microRNA (sometimes termed ‘seed’) appears to be one of the key determinants of target recognition. The reduction of target gene expression appears to be achieved by one or both of two mechanisms: inhibition of translation initiation or degradation of the mRNA.
Several resources provide microRNA target predictions based on sequence complementarity to target sites with emphasis on perfect base-pairing in the seed region and sequence conservation [TargetScan (3), PicTar (4), TargetRank (5)]. Other target predictions methods are based on calculations of mRNA secondary structure and energetically favorable hybridization between microRNA and target mRNA [RNAhybrid (6), STarMir (7)]. Additional information resources record experimentally validated microRNA targets [TarBase (8)], sequence annotation such as genomic organization, precursor sequences and literature citations [miRBase (9), miRGen (10), Argonaute (11), miRNAMap (12)], microRNA expression [smiRNAdb (13)] and miRNA transcriptional regulation (miRPromoter—unpublished data).
The microRNA.org site (http://www.microrna.org) is a resource for microRNA target predictions and microRNA expression that is widely used by the research community. We generated a new set of target predictions for human, mouse and rat microRNAs using the miRanda algorithm (14). The microRNA expression profiles were taken from a recent profiling study across ∼250 small RNA libraries collected from human, mouse and rat tissues and cell lines (13). A new graphical interface enables users to investigate predicted microRNA targets and their binding sites as well as microRNA expression in various tissues.
TARGET PREDICTIONS
Target predictions are performed using the miRanda algorithm (14,15) that computes optimal sequence complementarity between a set of mature microRNAs and a given mRNA using a weighted dynamic programming algorithm. The key extension to the Smith–Waterman algorithm is that the alignment score is a weighted sum of match and mismatch scores for base pairs (including G:U wobbles) and gap penalties. Weights are position-dependent and reflect the relative importance of the 5′ and 3′ regions in a finely adjustable way. The weight of each position can be optimized to reflect experimental facts and physical principles. In addition, miRanda uses an estimate of the free energy of formation of the microRNA:mRNA duplex [Vienna package (16)] as a secondary filter.
A natural consequence of the weighted alignment optimization is the inclusion of potential targets with some mismatches at the dominant 5′ end of the microRNA, but with otherwise good complementarity to the target gene. To reflect the importance of the 5′ region, base-pairing in positions 2–8 of the microRNA are given higher weights when computing the microRNA:mRNA alignment score. This approach, as designed into the original miRanda algorithm (14,15), is congruent with experimentally validated targets that do not contain perfect seed matches (17,18) and includes what other approaches have subsequently introduced as 3′-compensatory matches (19) or combinations of seed and 3′ match rules (20).
Sequence conservation at and near microRNA-binding sites is a strong indication of functional constraints in evolution and may reflect conserved RNA structure, accessibility of target sites, binding of helper proteins or ribonuclear complexes and other determinants. In principle, the notion of a conserved microRNA-binding site implies a conservation of microRNA:mRNA relationship (regulator and regulated), rather than just conservation of mRNA sequence. The conservation of this relationship depends on: (i) conservation of the mature microRNAs sequences, (ii) conservation of the relevant parts of the mRNA sequences and (iii) the presence of a homologous microRNA-binding site on the mRNA, which in general does not have to be in a similar position. In the current version of the algorithm we use a conservation filter, which is different than the one used previously but is equally informative (15). Specifically, to filter out less-conserved predicted target sites, we use the PhastCons conservation score, which measures the evolutionary conservation of sequence blocks across multiple vertebrates using a phylogenetic hidden Markov model (21). This filter does not separately compute details of the target match in the related organisms. In the limit of very strong conservation, target site details are very similar by definition. The filter does imply approximate conservation of target site position on the mRNA. As a default parameter, we use a PhastCons score cutoff of 0.57 to select target sites that are conserved in mammals.
DATA SOURCES AND IMPLEMENTATION
MicroRNA sequences are collected and routinely updated from MirBase(9) (current version 10.0). Mammalian 3′-UTR sequences from the rat (rno4), mouse (mm7) and human (hg18) genomes were downloaded from the UCSC genome browser (22). MicroRNA expression profiles were collected from a recently published comprehensive cloning and sequencing effort of 172 human, 64 mouse and 16 rat small RNA libraries extracted from major organs and cell types (13). Expression values represent the number of cloned mature microRNAs that were sequenced in each library and reported as normalized clone counts. Such clone counts are assumed to be monotonically related to cellular microRNA concentrations although this correlation may differ between microRNAs. In addition to precise measurements of expression levels, this comprehensive microRNA survey provided detailed information about the mature sequences, precursor forms, genomic organization and sequence conservation.
All the data were organized in a set of relational MySQL databases. The website implementation is based on Java, Struts, JSPs and Javascript as well as processing (http://processing.org/), an open-source graphics system that compiles into Java code. These applets allow the user to dynamically explore the data while reducing the amount of client–server interactions. Target predictions and miRNA expression values are freely available for download from the website, as well as the miRanda source code.
SCIENTIFIC USE CASES
The microRNA.org data resource is designed to address a number of common questions pertaining to microRNA regulation and expression. What are the predicted gene targets of a microRNA? What are the predicted microRNAs targeting a gene? What are the sequence positions of the microRNA:mRNA target sites? Which are the most abundant microRNAs in a specific tissue? In which tissues is a particular microRNA expressed? In Table 1, we outline a number of use case scenarios
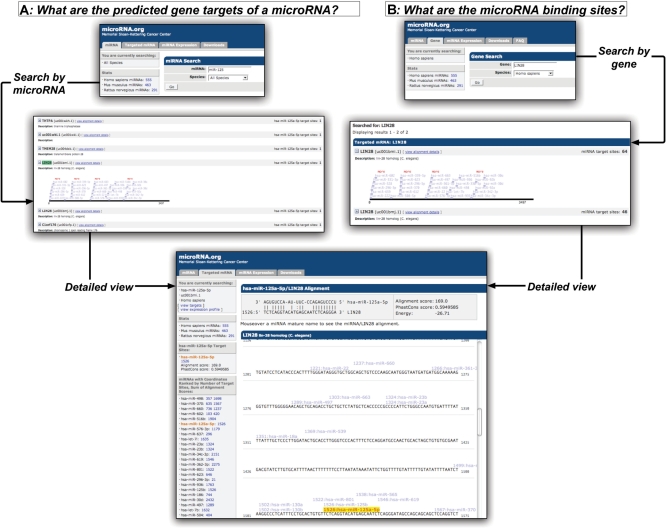
Figure 1.
Scientific use cases for exploring microRNA target predictions. In case A, users search for the ‘predicted target genes of a given microRNA’. In the gene list view, an expandable display shows a schematic view of the microRNA target sites on a selected target gene. A selected target gene is also hyperlinked to a detailed view of the microRNA-binding sites in the 3′-UTR, including exact nucleotide positions, microRNA:mRNA duplex, alignment score and conservation as well as the ranked list of the binding sites. Similarly, in case B, users can reach the detailed target-sites-on-sequence view by ‘querying by gene name’.
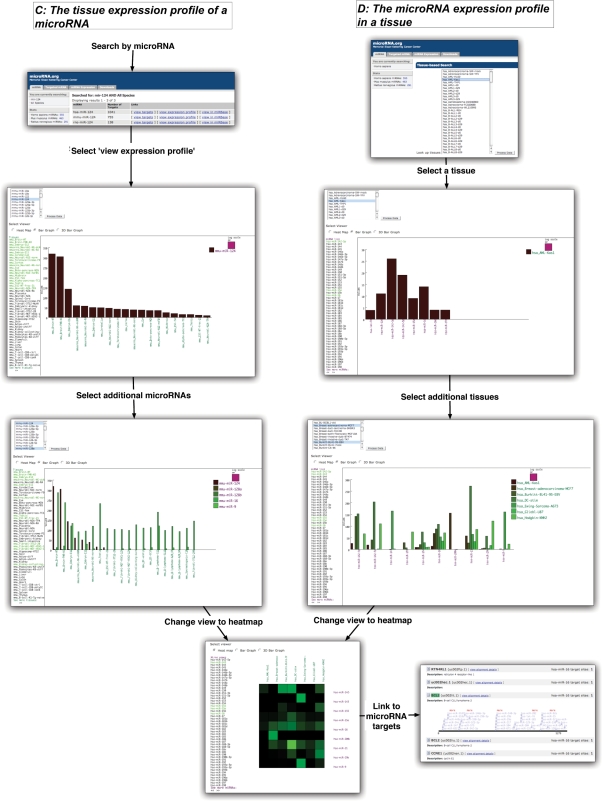
Figure 2.
Scientific use cases for exploring microRNA expression. In case C, users can select to view the ‘tissue expression profile of a microRNA’. Additional microRNAs can be incorporated into the graph from the list of microRNAs at the top of the view. Additional tissues can also be added or removed by selection from the list on the left. In the parallel instance D, users can select to ‘view the microRNA expression profile of a tissue (or a list of tissues)’. By analogous selections, users can incorporate additional microRNAs and tissues into the view from their respective lists. In both use cases, the data can be visualized as a heat map: vertical and horizontal axes represent tissues and microRNAs and colors represent microRNA expression levels. Additionally, in all views microRNAs are linked to their target view.
Table 1.
Scientific use case scenarios
| Cases | Question | Query process | Results |
|---|---|---|---|
| A | What are the target genes of a given microRNA? | Select the ‘miRNA’ tab and enter a full or partial microRNA name. From the returned list of microRNA names select the ‘view targets’ link for the microRNA of interest. | The list of target genes for this microRNA. Each gene is linked to a full mRNA sequence view with details of microRNA-binding sites (Figure 1). |
| B | What are the microRNAs targeting a given gene and what are their binding sites? | Select the ‘Targeted mRNA’ tab and enter the gene name, synonym or identifier (NCBI accession or RefSeq). From the list of matched genes select the desired gene. Entries with the same name are isoforms of the same gene, often with similar 3′-UTR. | A detailed view of the gene and its microRNA-binding sites. A ranked list of the microRNAs appears alongside the sequence view (Figure 1). |
| C | What is the tissue expression profile of a given microRNA [or set of microRNAs]? | Select the ‘miRNA’ tab and enter a full or partial microRNA name. From the returned list of microRNAs select the ‘view expression profile’ link for the microRNA of interest. | A graph of the expression levels in the top 20 tissues in which the microRNA is expressed. You can modify the graph by adding or removing tissues or microRNAs. The display can be toggled between a bar graph (2D), a composite bar graph (partial 3D) or a heat map (Figure 2). MicroRNAs are linked to their target view. |
| D | What is the microRNA expression profile in a given tissue [or set of tissues]? | Select the ‘miRNA Expression’ tab and from the returned list select the organism and tissues of interest. | A graph of the expression levels of the 20 top microRNAs expressed in the tissue. You can modify the graph by adding or removing tissues or microRNAs. The display can be toggled between a bar graph (2D), a composite bar graph (partial 3D) or a heat map (Figure 2). MicroRNAs are linked to their target view. |
FUTURE EXTENSIONS
The discovery process for new microRNAs and their functional roles is facilitated by computational efforts such as target predictions, predictions of microRNA genes and their transcriptional control, computational analysis of feedback regulation and the analysis of microRNA expression profiles. The microRNA.org database, now completely re-designed and re-engineered, has been a valuable resource for many computational and experimental studies since 2003. In future developments, we plan to include microRNA target predictions for Drosophila, Caenorhabditis elegans, Zebrafish and other organisms, as well as expression data from additional sequencing, array-based or bead-based profiling efforts, e.g. from cancer genomics projects.
Target and expression data can be linked in interesting ways. An obvious requirement for microRNA regulation is the concurrent expression of both a microRNA and its target genes. Ongoing profiling of microRNA expression will provide the necessary information to focus target predictions on a subset of microRNAs relevant to particular cell types and physiological or genetic conditions. Currently, users can link from the expression view to the microRNA target view. In future revisions, users will be able to restrict target predictions to a subset of microRNAs of interest in a particular biological context.
Particularly valuable information is provided by experiments that verify microRNA regulation and describe the role of microRNAs in specific pathways. We intend to incorporate data from recent efforts to capture experimental information in computable form, such as TarBase (8) and link microRNA target and expression information to pathway information resources, such at those available via pathwaycommons.org
We plan to continue to incorporate new details about microRNA regulation into the microRNA.org information resource, based on new experimental discoveries and improvements in prediction algorithms.
ACKNOWLEDGEMENTS
We thank Joanne Edington for systems support and Alex Lash for organizational support; Tom Tuschl, Pablo Landgraf, Rob Sheridan, Mihaela Zavolan and their co-authors for making available sequence and expression information; and, Anton Enright and Bino John for their earlier work on the miRanda algorithm. External funding was provided by a P01 grant from the US National Institutes of Health (NIGMS), Atlantic Philanthropies and the Alfred W. Bressler Scholars Endowment Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1.Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, et al. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J. Cell. Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 4.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 8.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. miRGen: a database for the study of animal microRNA genomic organization and function. Nucleic acids Res. 2007;35:D149–D155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shahi P, Loukianiouk S, Bohne-Lang A, Kenzelmann M, Kuffer S, Maertens S, Eils R, Grone HJ, Gretz N, et al. Argonaute – a database for gene regulation by mammalian microRNAs. Nucleic Acids Res. 2006;34:D115–D118. doi: 10.1093/nar/gkj093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu PW, Huang HD, Hsu SD, Lin LZ, Tsou AP, Tseng CP, Stadler PF, Washietl S, Hofacker IL. miRNA Map: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–D139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wuchty S, Fontana W, Hofacker IL, Schuster P. Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers. 1999;49:145–165. doi: 10.1002/(SICI)1097-0282(199902)49:2<145::AID-BIP4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 17.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 18.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]