Abstract
Pfam is a comprehensive collection of protein domains and families, represented as multiple sequence alignments and as profile hidden Markov models. The current release of Pfam (22.0) contains 9318 protein families. Pfam is now based not only on the UniProtKB sequence database, but also on NCBI GenPept and on sequences from selected metagenomics projects. Pfam is available on the web from the consortium members using a new, consistent and improved website design in the UK (http://pfam.sanger.ac.uk/), the USA (http://pfam.janelia.org/) and Sweden (http://pfam.sbc.su.se/), as well as from mirror sites in France (http://pfam.jouy.inra.fr/) and South Korea (http://pfam.ccbb.re.kr/).
INTRODUCTION
Pfam is designed to be a comprehensive and accurate collection of protein domains and families (1,2). Pfam families are divided into two categories, Pfam-A and Pfam-B. Each Pfam-A family consists of a curated seed alignment containing a small set of representative members of the family, profile hidden Markov models (profile HMMs) built from the seed alignment and an automatically generated full alignment which contains all detectable protein sequences belonging to the family, as defined by profile HMM searches of primary sequence databases. Pfam-B entries are automatically generated from the ProDom database (3), and are represented by a single alignment. The use of representative seed alignments for Pfam-A families allows efficient and sustainable manual curation of alignments and annotation, while the automatic generation of full alignments and Pfam-B clusters ensures that Pfam is a comprehensive classification of protein families that scales effectively with the growth of the sequence databases. Pfam data are freely accessible via the web and are available for download in a variety of forms (see availability section).
COVERAGE UPDATE
We quantify the completeness of the Pfam classification of sequence space using two measures of coverage. ‘Sequence coverage’ is the fraction of protein sequences listed in UniProtKB (4) that has at least one Pfam domain, whilst ‘residue coverage’ is the fraction of protein residues that fall within Pfam domains, as defined by the sub-sequences included in Pfam-A full alignments. The current Pfam release (version 22.0) contains a total of 9318 Pfam-A families, which cover 73.23% of sequences and 50.79% of residues found in UniProtKB version 9.7. Since the last Pfam publication (release 18.0), we have added 1335 families (a 17% increase) and maintained approximately the same coverage of UniProtKB, despite a 100% increase in the number of UniProtKB sequences.
Coverage of protein structures
The availability of 3D protein structures has been essential for finding distant evolutionary relationships and understanding protein function at the molecular level. Sequences with a known structure usually have a clear domain organization and ideally each domain should be represented in Pfam. We looked at a non-redundant set of sequences whose structures were deposited in the Protein Data Bank (PDB), and found more than 1000 structures, which did not have any corresponding Pfam-A domains. We also found that residue coverage of some PDB structures was <50%. In order to improve this situation, we built over 500 new Pfam-A families for PDB sequences and SCOP (Structural Classification of Proteins) database entries (6), which were not previously covered. As measured on the current database of non-redundant sequences of known structure (3 August 2007), our sequence and residue coverages are now 94.7% and 77.5%, respectively. We have developed a protocol to ensure that this level of coverage is maintained as the number of protein structures increases. As novel structures are identified through structural genomics programs so their sequences will be made a priority for curation into Pfam.
Expansion of Pfam clans
The concept of clans was introduced into Pfam in 2005 (2). Briefly, a clan is a collection of Pfam-A entries that are judged likely to be homologous. Clans are built manually, based on a wide variety of information sources: the primary literature, known structures, profile–profile comparisons and other databases such as SCOP (6). We try to represent the relationships between member families graphically by providing clan alignments. Clans thus define a simple hierarchical classification of Pfam entries, allowing better transfer of structural and/or functional information between families, and better predictions of function and structure for families of unknown function.
To infer relationships between families, we now use three different tools. In addition to the profile–profile comparison tool PRC (http://supfam.mrc-lmb.cam.ac.uk/PRC/), we now use another profile–profile comparisons tool HHsearch (7), and the simple comparison of outputs program, SCOOP (8). We use three different computational methods because each is sensitive to a slightly different set of relationships and, more importantly, the combination of the three tools reinforces independently detected relationships. Notably, SCOOP has allowed us to infer many novel relationships that were not detected using either of the profile–profile comparison tools. Pfam version 18.0 (the first major release of Pfam with clans) contained 172 clans, comprising 1181 Pfam-A families. As new families are added to Pfam and new relationships are discovered, we build new clans and add families to existing ones. In the current release of Pfam (version 22.0) there are now 283 clans, comprising a total of 1808 Pfam-A families, an increase of 53% since release 18.0. The proportion of Pfam domain hits that fall within a clan has increased from 31% in release 18.0 to 43% in release 22.0. This shows that many families in Pfam are related and that, to date, many of the largest Pfam-A families have been assigned into clans. We expect the clan classification to grow still further, since many automatically detected relationships still need to be manually verified.
IMPROVING ACCESS TO PFAM
Pfam website development
Although Pfam data have always been centrally maintained and curated, historically each member of the Pfam consortium has run a separate website to serve the same data. The three primary mirror sites are based in the UK, Sweden and the USA, with a further two recognized mirror sites in France and South Korea. Each of the primary consortium sites has tended to adopt a different look and feel and, although all sites have provided the same set of core services, each has also provided some additional tools and services that are unique to that particular site. This has lead to an entirely different user experience at each Pfam site, and has led to users’ confusion as to which site provides which services. The development of three main websites also caused a significant duplication of effort for the Pfam consortium.
A new Pfam website has been developed, with the goal of providing a single, unified website for Pfam data and services, that combines the best features of the separate sites in a single, common interface. In re-designing the website, we have been able not only to improve the navigation and architecture of the website itself, but also to design a more easily extensible and maintainable code-base for the future. This new code-base will be common to and developed by all members of the Pfam consortium. Furthermore, the new website code has been written with portability in mind and has been made publicly available, so that users may install and run the website locally if desired.
We have improved the organization and presentation of Pfam data. Everything related to, for example, a Pfam-A family, is collected into a single page, which is sub-divided into tab-panes that the user can easily switch between. Figure 1 shows a typical page for a Pfam-A family. We have similar tab-layout pages for data related to protein sequences, Pfam-B families, Pfam clans, proteome data from completed genomes and 3D protein structures. Each type of page represents a different route into the Pfam data, and each tabbed page provides links that allow the user to navigate easily between these different sections of Pfam. Additionally, users can browse lists of Pfam families or clans and can jump quickly between any type of entry in the site via a ‘jump to’ box found on most pages.
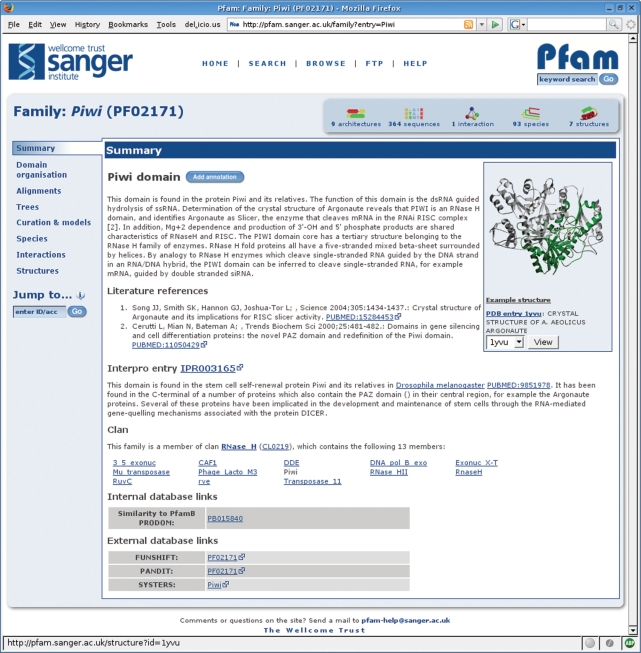
Figure 1.
The Pfam family page from the new website. This page shows the summary information for the Piwi domain. The tabs on the left allow users to browse the different types of associated information and beneath the tabs is the ‘jump to’ box, a tool which can direct the user to the page for any other entry in the site, given any type of accession or identifier. The panel at the top right gives a summary of the number of protein architectures, sequences, interactions, species and structures available. The same page layout and navigational tools are common to all of the different types of data in the Pfam website.
A common feature of every type of page is a summary box, providing the salient details of every entry in a single glance. The five summarized features of the entry are: the number of architectures associated with the entry; the number of protein sequences; the number of interactions [as determined by iPfam (9)]; the number of species; and the number of 3D structures. The exact meaning of each value is context-dependent, so that in the Pfam family page, for example, the structure icon shows the number of structures associated with that family, whilst in a protein sequence page the structure icon shows the number of structures, which map to that sequence. The link for each icon is also context-dependent, taking the user to the most appropriate section of the page for the icon clicked.
Previously, it has been difficult to search Pfam by species or taxonomic division. In addition to the species tree found on each family page, which provides a breakdown of the species found in that family, we have implemented a new taxonomy search tool. As with the taxonomy search tool in the old Pfam website, the new tool returns a list of Pfam domains that match a Boolean query expression. For example, the query ‘Caenorhabditis elegans AND NOT Homo sapiens’ will return all Pfam domains found in C. elegans, but are not found in H. sapiens. As well as being less error prone and significantly quicker than the version in the old Pfam website, the new taxonomy search tool also provides a feedback mechanism that suggests organism names as the user enters them. This reduces the likelihood of typographical or spelling errors in queries, since incorrectly entered species terms are immediately highlighted in the interface, as well as providing an insight into the organisms that are found in the database.
A commonly requested capability for the Pfam site is the ability to find Pfam domains that are unique to a given taxonomic division or species. This feature is now available. For example, searching for unique ‘Metazoa’ families returns a list of domains that are found only in Metazoans will be returned.
In addition to the standard features of the old Pfam websites, such as search tools for quickly finding Pfam domains on a protein sequence or for locating sequences with a specified domain architecture, we have also introduced several new features in the new site, many of which use the Distributed Annotation System (DAS) (10) to aggregate multiple data sources in a single display.
The Distributed Annotation System
We have improved access to Pfam by providing data through the DAS. DAS is a system for disseminating annotations and alignments of DNA or protein sequences through a simple, web-based protocol. Three types of Pfam data are now available via DAS (11): domain annotations for both Pfam-A and Pfam-B families; sequence features such as active sites (12) and transmembrane region predictions; and seed and full alignments for Pfam-A families. The availability of Pfam data via DAS enables users to access specific parts of the database as a web service, without the need to download and install it in its entirety.
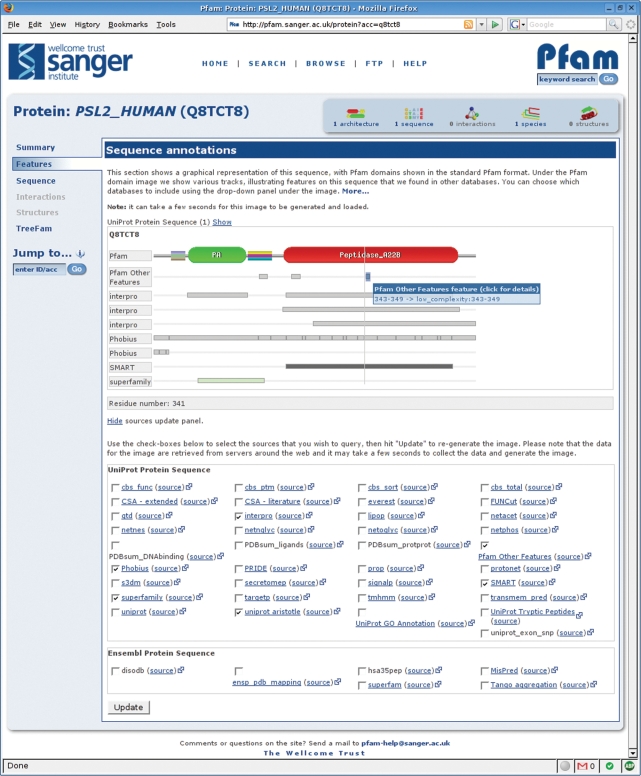
We have also been able to incorporate other data sources that are accessible through DAS, in order to enrich our own display of Pfam data. One feature of the new website is a DAS-based viewer for sequence annotations (Figure 2). This allows the user to view annotations of protein sequences from a wide range of third-party databases alongside information from Pfam itself. The viewer presents the standard Pfam domain structure image, showing the arrangement of Pfam domains on the sequence in question, and allows users to add or hide annotations from any of the available DAS sources. As the user moves their mouse over each feature, a tool-tip gives detailed information about it. If provided by the external DAS source, a link to further information is also given.
Figure 2.
The Pfam protein sequence page, showing the DAS annotation viewer. Various tracks can be selected using the check boxes beneath the annotation view, allowing the user to view features derived from any of the listed DAS sources. For instance, in the example displayed, the membrane topology calculated by Phobius (18) can be viewed alongside the Pfam domain annotations and those from a variety of different domain databases. The figure shows a tool-tip for one feature, which gives the details of the annotation and, in this case, highlights the link to further information.
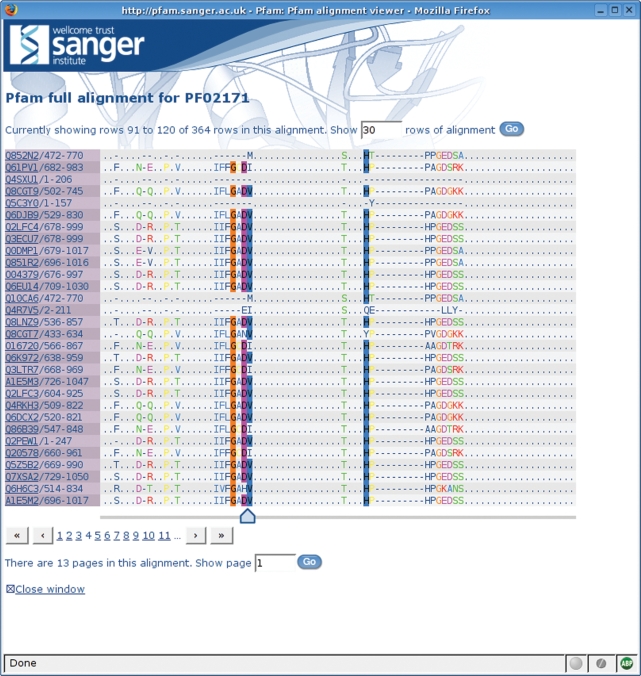
Another use of DAS within the new website is in the Pfam sequence alignment viewer. Pfam provides two alignments for every family: the seed alignment is a manually curated alignment of related sequences and generally contains a relatively small number of sequences; the full alignment is generated by searching the sequence database using the HMM for the family and may contain a very large number of sequences (the largest alignment, that of GP120, currently contains over 68 000 sequences). Historically, it has been difficult, if not impossible, to view the largest sequence alignments in a web browser, due simply to the size of the resulting web page. We have implemented a DAS-based sequence alignment viewer (shown in Figure 3) that is able to present even the largest alignments in manageable portions, by retrieving only the required section of the alignment and rendering it as HTML. This allows the user to scroll through wide alignments (those with long sequences) or to page through long alignments (those with a large number of sequences), without having to load the entire alignment into their browser. Alignments are coloured according to a pre-calculated consensus sequence, which is also retrieved via DAS, and in this way even alignment fragments can be marked-up using the properties of the whole alignment.
Figure 3.
The DAS-based Pfam sequence alignment viewer. A portion of the Piwi domain full alignment is shown with residue conservation highlighted in a ClustalX (19) style colour scheme.
WIDENING THE SCOPE OF PFAM ANNOTATIONS
NCBI GenPept sequences
The two main public repositories of protein sequence data are UniProtKB (4) and the GenPept database from NCBI (13). These two resources are independent of each other and consequently contain two separate, though often overlapping, sets of sequences, which are referred to by entirely separate sets of accessions. Historically, Pfam has been based on a sequence database termed ‘pfamseq’, which is a frozen version of the UniProtKB database that we update at each major Pfam release. This has caused problems for users wanting to retrieve Pfam data for a sequence for which they have only a GenPept accession or the NCBI GI number.
To make Pfam sequence annotation accessible via both UniProtKB and GenPept accessions, we now provide Pfam domain assignments for members of both sequence databases, each as a separate section of Pfam. Where possible, we transfer the annotation from UniProtKB sequences to the equivalent sequence in GenPept, by using the EMBL/GenBank cross-references (13,14) in the UniProtKB entry and ensuring that the CRC64 checksums of the sequences from the two databases are the same. For example, of the five listed protein identifiers cross-referenced by the UniProtKB accession P51003 (AAH00927.1, AAH36014.1, CAD62628.1, CAD66560.1, CAD61935.1), only one GenPept protein (AAH36014.1) has the same CRC64 checksum as the UniProtKB entry. As a quality control procedure, we make use of the UniProtKB/Swiss-Prot mapping provided by GenPept to perform the reverse mapping. Overall, this mapping procedure provides a UniProtKB equivalent for around 75% of sequences in GenPept. We search the remaining GenPept sequences against the library of Pfam HMMs and use the pre-defined, curated thresholds to determine which sequences should be included in the full alignment for a family. Two or more families belonging to the same clan may match the same sequence region. We resolve such overlaps using the same method as we use when resolving overlapping matches between clan families that have been searched against the UniProtKB sequence database (2). This ensures that there are no overlapping sequences between families that belong to the same clan.
The Pfam domain annotations and alignments for GenPept (release 158) are available for download in a flat-file format (Pfam-A.full.ncbi), as an ASCII representation of the domains matches on each sequence (similar to the swisspfam file) and in the Pfam MySQL database. We also provide access to this data via the websites, where Genbank identifiers (GI numbers) or GenPept protein identifiers can be entered into the ‘jump to’ box. Searching in this way will take users to a page that is similar to the protein pages produced for UniProtKB sequences. One caveat when using this data is that we do not resolve overlaps between domains for GenPept sequences. However, since we curate Pfam-A domain thresholds in a conservative manner to ensure high specificity (at the expense of some sensitivity), we expect the number of domain overlaps for the GenPept data to be low. Furthermore, three-quarters of the sequences in GenPept are identical to a UniProtKB entry, which are guaranteed to be non-overlapping.
Metagenomic samples
Current microbiological culturing techniques are inadequate for studying the vast majority of microorganisms (15). Consequently, many organisms remain under-represented in the main sequence databases. An emerging field in biology is metagenomics, the analysis of genomic material from environmental samples. Recently, with the advent of better sequencing technologies, large samples from environments such as the sea have been sequenced directly, thereby avoiding the need for culturing. Sequencing using this approach gives rise to many sequences from a diverse set of organisms, albeit at low read coverage and with no knowledge of the source organisms. In addition, when compared with proteins in UniProtKB, the sequences from metagenomic samples are more fragmentary.
Within Pfam we have collected together several available metagenomic datasets and amalgamated them into a single sequence collection (listed on the ftp site), which we have termed ‘metaseq’. Currently, the only centralized public repository for such sequences that we are aware of is the UniProt Metagenomic and Environmental Sequence database (UniMes). However, UniMes currently only contains data from the Global Ocean Sampling (GOS) Expedition (16), the largest and most publicized of such environmental sampling projects. When the UniMes database becomes more comprehensive, we will use this as the underlying sequence database.
There are currently more than 6.6 million sequences in the ‘metaseq’ database, making it significantly larger than the current version of ‘pfamseq’, which currently contains around 4.5 million sequences. We have searched the sequences in ‘metaseq’ against the library of Pfam HMMs. All sequence regions that score above the pre-defined curated thresholds have been recorded, and for each family, the significant matches aligned, in the same manner as we generate our full alignments. These domain annotations and alignments are available both in flat-file formats (as with the NCBI GenPept dataset) and in the MySQL database. The metagenomics domain annotations can also be retrieved via the website. Similar to the NCBI dataset, ‘metaseq’ accessions and identifiers can be used to retrieve a graphical representation of the sequence and Pfam domains (if any have been found). As with the GenPept data, we have not resolved any overlaps, but, unlike the situation with the GenPept data, we have not ‘competed’ the overlapping sequence hits for families within clans, which means that there will inevitably be some overlapping hits between families that belong to the same clan.
The metagenomics dataset contains many novel protein sequences, which are currently unannotated. This section within Pfam enables the community to assess our current understanding of the domain composition found in such environmental datasets. Uniquely, users will also be able to access domain alignments that can be compared to those historically found in Pfam. It was estimated that there are over 1000 new protein families that are not currently represented by Pfam in the GOS dataset alone (17). Thus, this dataset will provide a potential source of new Pfam families and/or allow verification of families where there are few representatives found in UniProtKB.
SUMMARY
In the last 2 years the Pfam database has continued to grow, improving both coverage and quality of families. In particular, we have widened the scope of Pfam to include sequences from the GenPept database, as well as providing matches to new metagenomics sequence data. We have also developed a new website that provides a unified view for the primary Pfam consortium sites. Pfam has been curating protein families for over 10 years, but there is still much to be done to provide a complete and accurate classification of proteins.
AVAILABILITY
Pfam data can be downloaded directly from the WTSI FTP site (ftp://ftp.sanger.ac.uk/pub/databases/Pfam), either as flat files or in the form of MySQL table dumps. You can visit new Pfam websites at WTSI (http://pfam.sanger.ac.uk/), Stockholm Bioinformatics Center (http://pfam.sbc.su.se/) and Janelia Farm (http://pfam.janelia.org/). The source code for the website can be retrieved by CVS from the WTSI CVS repository, that can be browsed at http://cvs.sanger.ac.uk/cgi-bin/viewcvs.cgi/?root=PfamWeb. Instructions for downloading the code directly from CVS are available at http://cvs.sanger.ac.uk/cvs.users.shtml.
ACKNOWLEDGEMENTS
The authors would like to thank Roger Pettett and Jody Clements for useful discussions on the design and implementation of the new website. We are grateful for the infrastructure support provided by Guy Coates, Tim Cutts and Andy Bryant at Wellcome Trust Sanger Institute (WTSI). Finally, we would to like to thank all of the users of Pfam who have submitted new families and/or annotation updates for existing entries. R.D.F., J.T., J.M., P.C.C., S.J.S., H.-R.H. and A.B. are funded by The Wellcome Trust, R.D.F. and J.M. were funded partly by an MRC (UK) E-science grant (G0100305). S.R.E. and G.C. are supported by the Howard Hughes Medical Institute. K.F. and E.L.L.S. are funded by Stockholm University, Royal Institute of Technology and the Swedish Natural Sciences Research Council. Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sonnhammer ELL, Eddy SR, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 2.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bru C, Courcelle E, Carrere S, Beausse Y, Dalmar S, Kahn D. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33:D212–D215. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. [Google Scholar]
- 5.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 8.Bateman A, Finn RD. SCOOP: a simple method for identification of novel protein superfamily relationships. Bioinformatics. 2007;23:809–814. doi: 10.1093/bioinformatics/btm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finn RD, Marshall M, Bateman A. iPfam: visualization of protein–protein interactions in PDB at domain and amino acid resolutions. Bioinformatics. 2005;21:410–412. doi: 10.1093/bioinformatics/bti011. [DOI] [PubMed] [Google Scholar]
- 10.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finn RD, Stalker JW, Jackson DK, Kulesha E, Clements J, Pettett R. ProServer: a simple, extensible Perl DAS server. Bioinformatics. 2007;23:1568–1570. doi: 10.1093/bioinformatics/btl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mistry J, Bateman A, Finn RD. Predicting active site residue annotations in the Pfam Database. BMC Bioinformatics. 2007;8:298. doi: 10.1186/1471-2105-8-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulikova T, Akhtar R, Aldebert P, Althorpe N, Andersson M, Baldwin A, Bates K, Bhattacharyya S, Bower L, et al. EMBL Nucleotide Sequence Database in 2006. Nucleic Acids Res. 2007;35:D16–D20. doi: 10.1093/nar/gkl913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schloss PD, Handelsman J. Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol. 2005;6:229. doi: 10.1186/gb-2005-6-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, et al. ClustalW and ClustalX version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]