Abstract
Binding MOAD (Mother of All Databases) is a database of 9836 protein–ligand crystal structures. All biologically relevant ligands are annotated, and experimental binding-affinity data is reported when available. Binding MOAD has almost doubled in size since it was originally introduced in 2004, demonstrating steady growth with each annual update. Several technologies, such as natural language processing, help drive this constant expansion. Along with increasing data, Binding MOAD has improved usability. The website now showcases a faster, more featured viewer to examine the protein–ligand structures. Ligands have additional chemical data, allowing for cheminformatics mining. Lastly, logins are no longer necessary, and Binding MOAD is freely available to all at http://www.BindingMOAD.org.
INTRODUCTION
The field of structure-based drug design relies on high-quality databases of protein–ligand structures to develop the best computational tools. There are several available, including but not limited to Binding MOAD (1), PDBbind (2), LPDB (3), Relibase (4), BindingDB (5), PDBLig (6), MSDsite (7), eF-Site (8), PDB-Ligand (9), SuperLigands (10), PLD (11), HET-PDB (12), sc-PDB (13), PDBsite (14), Ligand Depot (15), AffinDB (16) and KiBank (17). Each database has a unique focus and incorporates different data content, chemical structures, and/or analysis tools.
Our development of Binding MOAD focuses on providing the largest collection of high-quality, protein–ligand complexes. Each structure is hand curated by reading the crystallography paper which presents the structure in the literature; this is used to validate ligands and acquire binding affinities. Binding MOAD contains all appropriate protein–ligand complexes: protein–ligand, protein-cofactor and protein–ligand-cofactor. It also presents complexes even when no binding data is currently available. This makes it the largest database of its type. Here, we discuss the latest update to the Binding MOAD database, outlining improved access, the addition of new data and the incorporation of new tools.
BINDING MOAD COMPOSITION
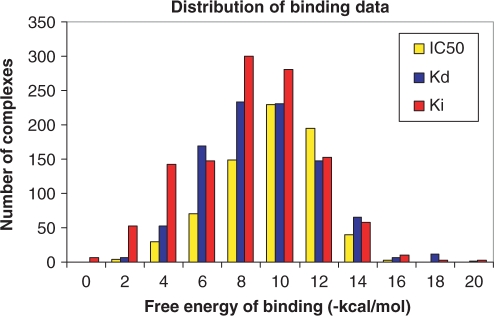
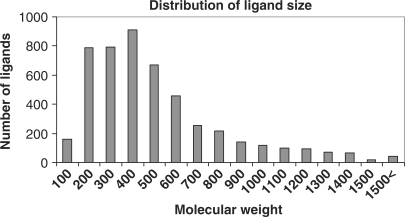
Binding MOAD is constructed with a top-down approach, starting with all entries in the PDB (18) and eliminating structures which are inappropriate. This is more efficient than a ground-up approach of reading the literature as a whole to identify appropriate complexes. Each entry in Binding MOAD must have resolution better than 2.5 Å, and each entry must contain a valid ligand. Valid ligands are biologically relevant small molecules and can include agonists, antagonists, cofactors, inhibitors, allosteric regulators, enzymatic products, etc. Covalently attached molecules (covalent inhibitors or posttranslational modifications to the protein) are not considered valid ligands. The focus is proteins binding small molecules, so peptides larger than 10 amino acids and chains of greater than four nucleic acids are not considered valid ligands. Many small molecules present in a crystal structure are not considered biologically relevant because they are part of the crystallization matrix and an artifact of the protein in an artificial condensed phase. Such molecules include solvents, buffers, detergents and salts, but care must be taken because some small molecules are valid ligands in some structures but additives in others. Examples of such are sugars, membrane components, small organic molecules (e.g. toluene) and metabolites (e.g. citrate). Figures 1 and 2 illustrate the wide distribution of data available in Binding MOAD, in terms of binding affinity and size, respectively.
Figure 1.
Distribution of binding affinities in Binding MOAD. Data is labeled as Kd (blue), IC50 (yellow) and Ki (red). For this figure, binding affinities were simply converted to free energies by RT × ln(affinity). While this conversion not strictly appropriate for Ki or IC50, it provides a comparison for the reader.
Figure 2.
Distribution of the sizes of 5074 unique ligands in Binding MOAD. The largest ligands are peptides, short oligonucleotides and complex sugars.
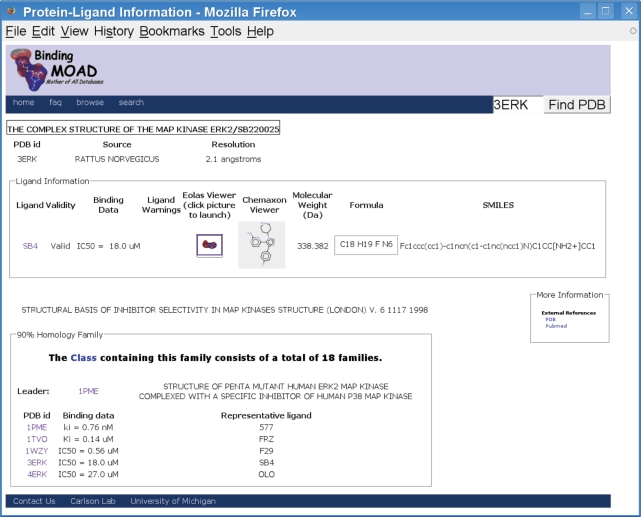
In Binding MOAD, proteins are grouped into families of 90% sequence identity. By choosing one representative of each family (the ligand with the best affinity), we can create a non-redundant set which removes any skew resulting from proteins that are heavily represented in the PDB (18). Protein families are also grouped by function using Enzyme Classification numbers and our own non-enzymatic classifications. At the bottom of the data page for each complex in Binding MOAD, the entire protein family is reported and a link is provided to view all the data for that functional class (Figure 3). This allows a user to start at a particular complex important to his/her research, and from there, jump to other related structures.
Figure 3.
Screenshot of the data page for 3ERK, showing the additional ligand data and the connectivity to proteins with similar structure and function.
GROWTH
Since its introduction in 2004, Binding MOAD has regularly expanded its collection with new data. Originally it contained 5331 protein–ligand complexes, and it has increased by almost 1500 each year, growing to 6638 in 2005, 8250 in 2006 and 9836 with the latest update. This steady growth mirrors the growth of the PDB; each year's update has consistently shown that one-fourth of the PDB structures meet our criteria for inclusion in Binding MOAD. Of the 9836 entries in the current version, 2950 (∼30%) have binding data curated from literature. It contains 3153 protein families and 5074 unique ligands.
As noted earlier, each crystallography paper is read to classify the ligands and extract affinity data for the ligand. Thus, adding new data to Binding MOAD involves reading a tremendous number of journal articles for manual annotation. After reading ∼10 000 papers, we have turned to automated methods to facilitate the process! A workflow tool, Binding Unstructured Data Analysis (BUDA), has been developed employing natural language processing (NLP) to evaluate and organize the papers for each update cycle. It identifies key sentences and phrases in papers and uses a weighted-scoring algorithm to rank the likelihood that the key sentences and phrases contain binding data. The NLP portion of BUDA is built upon the General Architecture for Text Engineering (GATE) framework (gate.ac.uk). The workflow portion of BUDA is used by the curators to organize the data for the annotation process. From the workflow interface, the curators can sort the articles by their weighted scores, review texts with highlights noting key phrases or sentences, and update the data into Binding MOAD. One of the key features is the ability to score a paper as lacking affinity data; it is a significant time-saving measure, rather than reading the paper in vain. While NLP can be used to speed up and guide the literature step, we unfortunately cannot use NLP to automatically extract the desired information. Many papers give affinity information for related systems when such information is unavailable for the exact complex in the crystal structure (e.g. affinities for wild-type protein are reported but the structures are all mutants or only the range of affinities is given for an entire inhibitor class). The data in Binding MOAD is for precisely the protein–ligand pair in the crystal structure, so data must be specific for that ligand bound to that exact protein. This evaluation must be made by hand.
AVAILABILITY
We have recently removed the need for users to login, and data is now freely accessible to all private companies, non-profits and foreign institutions. A comma-separated values (CSV) file is available to obtain the binding data and ligand information, organized by protein class and family. The CSV format was chosen for wide portability. Structures are also available as biounit files.
PLATFORM
Binding MOAD is built on proven technologies. The database is based on the Java 2 Platform, Enterprise Edition (J2EE), using an open-source JBoss Application Server, Enterprise JavaBeans (EJB) and a MySQL database backend. These tools provide a standards-compliant, easy-to-use website that unifies the presentation of structural, chemical and binding data in one simple format. In particular, the structure of the database allows us to expand the features and add new data easily.
RECENT IMPROVEMENTS
A screenshot of the modified layout for a data page in Binding MOAD is shown in Figure 3. New data has been incorporated about the valid ligands in each structure, including interactive 2D pictures, chemical formulae and the corresponding SMILES strings. As before, all ligands are noted as valid or invalid. When a hetgroup is considered part of the protein (glycosylation, catalytic metal, HEME group, etc.), it is not listed on the data page.
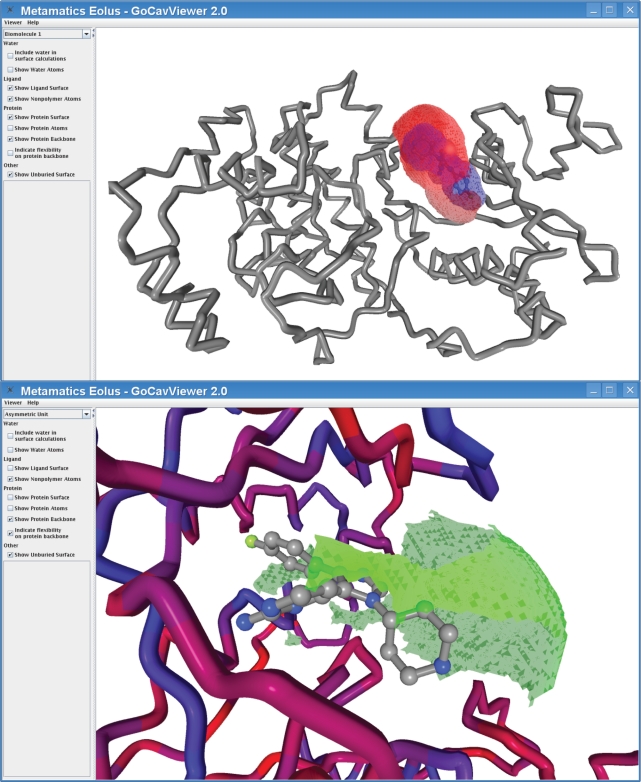
The greatest improvement comes as a new tool for viewing the complex in 3D. The GoCavViewer has been replaced by the EolasViewer; screenshot of the viewer is shown in Figure 4. As before, the viewer is capable of displaying the ligand pocket using both ball-stick and surface representations; the surfaces come from our code GoCAV which was specifically developed for Binding MOAD (19). However, EolasViewer incorporates significant improvements in the areas of performance, visual quality and back-end flexibility for future application development efforts.
Figure 4.
EolasViewer for 3ERK. The SB4 ligand is shown in ball in stick inside the pocket. The surfaces shown are the ligand surface in blue, the binding site in red and the solvent-exposed regions of the binding site are in green. (Top) The protein backbone is shown as a gray ribbon, and in the close-up (Bottom), the backbone is colored by B-factors.
The new viewer is built using the Eolus platform from Metamatics. Like its predecessor, the Eolus-based viewer is built using a Java framework, and the Binding MOAD website deploys it as a WebStart application. Eolus uses Jogl (Java Bindings for OpenGL) to fully utilize the 3D acceleration features available in nearly all modern computers. These two technologies, Java WebStart and OpenGL, provide nearly hands-free deployment of the software, together with state-of-the-art performance and visual quality. It takes advantage of rendering algorithms and OpenGL Shader Language (GLSL) to provide enhanced representation styles. The surface representation has been expanded to a fully transparent polygon surface. The proteins are rendered as ribbons by default, and the entire protein can now be rendered either as ribbon or ball-stick (the GoCavViewer was limited to displaying only residues that comprised the binding site). Finally, Eolus is a platform for structural biology, being developed in conjunction with this and other tools.
FUTURE DIRECTIONS
The data is currently organized with respect to protein structure and function, but we will expand the organization of the ligands by their chemical nature. At this time, ligands are annotated by their 3-letter HET codes, but full names will soon be added. A single-click search links all structures that contain the same molecule, but that is the extent of cross-linking by ligand data. We are adding similarity-based searches for the ligands. This effort will incorporate the new remediated ligand data released by the PDB consortium, and we look to cross-link our information with other major databases that focus on proteins and ligands. Furthermore, we look to use our text-mining tools to extend our search for affinity data beyond the crystallography literature. Lastly, Binding MOAD adds data once a year, but we look to make this a semi-annual event, given the success with NLP. Such NLP-based, text-mining approaches can be readily applied to other bioinformatic projects. This technology can be used to extract a wide variety of data—not just binding information—from the huge body of literature available today.
ACKNOWLEDGEMENTS
The authors wish to thank Steve Spronk for helpful discussion. We thank ChemAxon (www.chemaxom.com) for making web-based tools accessible to non-profit websites. This work was originally funded by a Young Investigator Award to H.A.C. from the Arnold and Mabel Beckman Foundation. It is currently funded by a CAREER Award to H.A.C. from the National Science Foundation (MCB 0546073). R.D.S. thanks the Molecular Biophysics Training Program (funded by the National Institutes of Health, GM 008270). N.A.K. thanks the Training Program in Bioinformatics (funded by the National Institutes of Health, GM 070449). Funding to pay the Open Access publication charges for this article was provided by the HAC's NSF grant (MCB 0546073).
Conflict of interest statement. None declared.
REFERENCES
- 1.Hu L, Benson ML, Smith RD, Lerner MG, Carlson HA. Binding MOAD (Mother Of All Databases) Prot. Struct. Func. Bioinf. 2005;60:333–340. doi: 10.1002/prot.20512. [DOI] [PubMed] [Google Scholar]
- 2.Wang R, Fang X, Lu Y, Yang CY, Wang S. The PDBbind Database: methodologies and updates. J. Med. Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 3.Roche O, Kiyama R, Brooks CL. Ligand-protein database: linking protein-ligand complex structures to binding data. J. Med. Chem. 2001;44:3592–3598. doi: 10.1021/jm000467k. [DOI] [PubMed] [Google Scholar]
- 4.Hendlich M, Bergner A, Gunther J, Klebe G. Relibase: design and development of a database for comprehensive analysis of protein-ligand interactions. J. Mol. Biol. 2003;326:607–620. doi: 10.1016/s0022-2836(02)01408-0. [DOI] [PubMed] [Google Scholar]
- 5.Liu T, Lin Y, Wen X, Jorrisen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalk AJ, Worth CL, Overington JP, Chan AWE. PDBLIG: classification of small molecular protein binding in the Protein Data Bank. J. Med. Chem. 2004;47:3807–3816. doi: 10.1021/jm040804f. [DOI] [PubMed] [Google Scholar]
- 7.Golovin A, Dimitropoulos D, Oldfield T, Rachedi A, Henrick K. MSDsite: a database search and retrieval system for the analysis and viewing of bound ligands and active sites. Prot. Struct. Func. Bioinf. 2005;58:190–199. doi: 10.1002/prot.20288. [DOI] [PubMed] [Google Scholar]
- 8.Kinoshita K, Furui J, Nakamura H. Identification of protein functions from a molecular surface database, eF-site. J. Struct. Funct. Genomics. 2002;2:9–22. doi: 10.1023/a:1011318527094. [DOI] [PubMed] [Google Scholar]
- 9.Shin J-M, Cho D-H. PDB-ligand: a ligand database based on PDB for the automated and customized classification of ligand-binding structures. Nucleic Acids Res. 2005;33:D238–D241. doi: 10.1093/nar/gki059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michalsky E, Dunkel M, Goede A, Preissner R. SuperLigands - a database of ligand structures derived from the Protein Data Bank. BMC Bioinformatics. 2005;6:122. doi: 10.1186/1471-2105-6-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puvanendrampillai D, Mitchell JBO. Protein Ligand Database (PLD): additional understanding of the nature and specificity of protein-ligand complexes. Bioinformatics. 2003;19:1856–1857. doi: 10.1093/bioinformatics/btg243. [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi A, Iida K, Matsui N, Tomoda S, Yura K, Go M. Het-PDB Navi.: a database for protein-small molecule interactions. J. Biochem. 2004;135:79–84. doi: 10.1093/jb/mvh009. [DOI] [PubMed] [Google Scholar]
- 13.Kellenberger E, Muller P, Schalon C, Bret G, Foata N, Rognan D. sc-PDB: an annotated database of druggable binding sites from the Protein Data Bank. J. Chem. Inf. Model. 2006;46:717–727. doi: 10.1021/ci050372x. [DOI] [PubMed] [Google Scholar]
- 14.Ivanisenko VA, Pintus SS, Grigorovich DA, Kolchanov NA. PDBSite: a database of the 3D structure of protein functional sites. Nucleic Acids Res. 2005;33:D183–D187. doi: 10.1093/nar/gki105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Z, Chen L, Maddula H, Akcan O, Oughtred R, Berman HM, Westbrook J. Ligand Depot: a data warehouse for ligands bound to macromolecules. Bioinformatics. 2004;20:2153–2155. doi: 10.1093/bioinformatics/bth214. [DOI] [PubMed] [Google Scholar]
- 16.Block P, Sotriffer CA, Dramburg I, Klebe G. AffinDB: a freely accessible database of affinities for protein-ligand complexes from the PDB. Nucleic Acids Res. 2006;34:D522–D526. doi: 10.1093/nar/gkj039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Aizawa M, Amari S, Iwasawa Y, Nakano T, Nakata K. Development of KiBank, a database supporting structure-based drug design. Comput. Biol. Chem. 2004;28:401–407. doi: 10.1016/j.compbiolchem.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith RD, Hu L, Falkner JA, Benson ML, Nerothin JP, Carlson HA. Exploring protein-ligand recognition with Binding MOAD. J. Mol. Graph. Model. 2006;24:414–425. doi: 10.1016/j.jmgm.2005.08.002. [DOI] [PubMed] [Google Scholar]