Abstract
The UCSD-Nature Signaling Gateway Molecule Pages (http://www.signaling-gateway.org/molecule) provides essential information on more than 3800 mammalian proteins involved in cellular signaling. The Molecule Pages contain expert-authored and peer-reviewed information based on the published literature, complemented by regularly updated information derived from public data source references and sequence analysis. The expert-authored data includes both a full-text review about the molecule, with citations, and highly structured data for bioinformatics interrogation, including information on protein interactions and states, transitions between states and protein function. The expert-authored pages are anonymously peer reviewed by the Nature Publishing Group. The Molecule Pages data is present in an object-relational database format and is freely accessible to the authors, the reviewers and the public from a web browser that serves as a presentation layer. The Molecule Pages are supported by several applications that along with the database and the interfaces form a multi-tier architecture. The Molecule Pages and the Signaling Gateway are routinely accessed by a very large research community.
INTRODUCTION
The UCSD-Nature Signaling Gateway (http://www.signaling-gateway.org) is a collaboration between the University of California, San Diego and the Nature Publishing Group (NPG), designed to facilitate navigation of the complex world of research into cellular signaling. The Signaling Gateway is made up of three components: the Molecule Pages (described in this study), the Signaling Update and the Data Center. The Signaling Update is published weekly by the NPG to provide topical and timely information about progress in signal transduction research. The Signaling Gateway was formerly sponsored by the Alliance for Cellular Signaling (AfCS) (1,2), which performed comprehensive experimental analyses of selected signaling systems. The Data Center section of the web site contains all the data generated by the AfCS during the period Signaling Gateway was part of the AfCS.
The Signaling Gateway Molecule Pages (SGMP) database provides essential information on more than 3800 proteins involved in cellular signaling in mammals, with each protein having its own Molecule Page. Molecule Page information is presented in two categories: author-entered data and automated data. Author-entered data contain expert-authored and peer-reviewed information based on published literature, with both review style free text with citations, and highly structured data for bioinformatic interrogation. The information is linked to appropriate journal citations, and covers areas such as protein interactions and states, transitions between states and protein function. The automated data information is a collection of public bioinformatic data source links and sequence analysis results, derived from the sequence and data record used to define the Molecule Page. The SGMP base organism is Mus musculus (because of the mouse-centric focus of the AfCS), though much of the information in the SGMP is derived from homologous proteins in other species, such as Homo sapiens.
Once the author-entered information has gone through a peer review process, the Molecule Page is published. The published Molecule Pages are citable and to date NPG has published entries for 365 proteins, with nearly 130 submitted Molecule Pages currently in the review system, and 350 Molecule Pages in author preparation. New published Molecule Pages are promoted through the Signaling Update website pages, e-alerts and linkouts from NPG content.
DATABASE CONTENT
The SGMP is a complicated online annotation and publishing system, containing three major subcomponents: (i) online pathway curation (author-entered data); (ii) online peer review and (iii) public repository data acquisition and display (automated data). The peer review information and the pre-published author-entered information for a given Molecule Page are only visible to the author, selected reviewers and the editorial staff, and are invisible to all other users. The automated data for each Molecule Page is visible to all users.
Each Molecule Page is assigned a specific protein sequence, a name, a list of synonyms and a specific protein function category (based on ‘best fit’). That information is used to generate the properties of the sequence such as molecular weight, and all the automated data associated with the sequence. A combination of database links and computational methods are used to find the related database records and the parameters of computational matches (e.g. a domain region). This information is displayed in the ‘Protein Overview’ section of the Molecule Page, which is the landing page for unpublished Molecule Pages.
Author-entered data
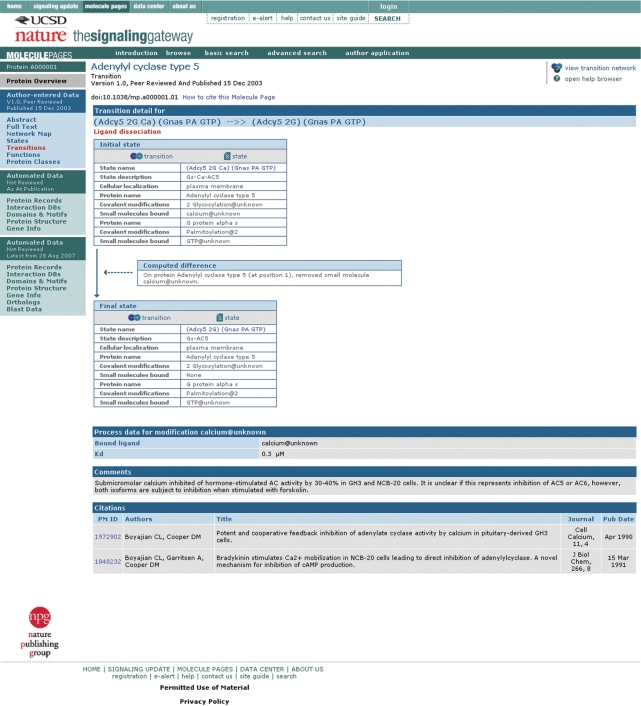
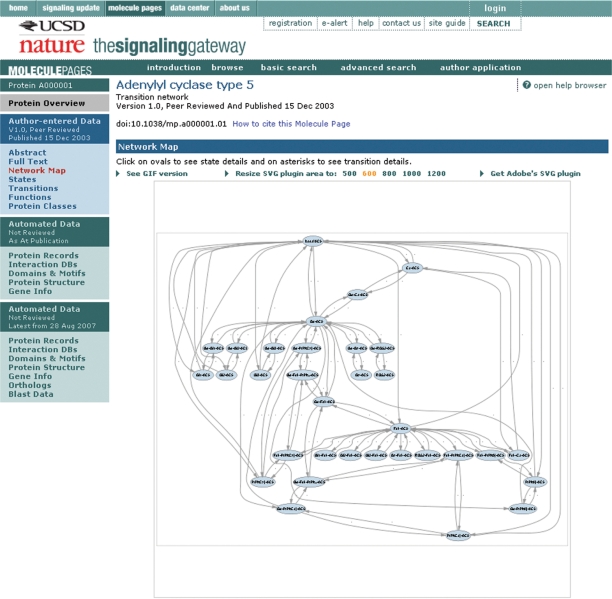
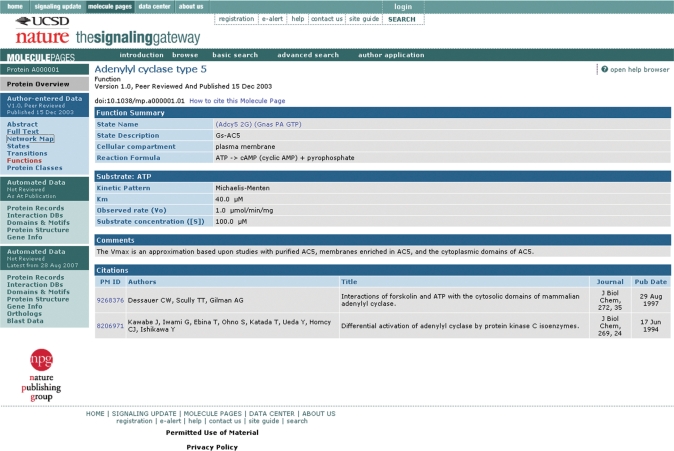
To illustrate the depth of the author-entered data, we choose Adenylyl cyclase type 5 (SGMP ID A000001). Because this is a published Molecule Page, the user first arrives at the ‘Abstract’ section, which gives information on the author (Carmen W. Dessauer), gives a summary of the role of Adenylyl cyclase type 5, lists the names and synonyms provided by the author and the editors, indicates that A000001 molecule has 32 enzyme functions, exists in 33 states, has 96 transitions between these states, and shows a miniature version of the network map of these transitions. The ‘Full Text’ section contains a textual description—with published references—of protein function, regulation, interactions, subcellular localization, expression, phenotypes, splice variants and antibodies. The ‘States’ section lists each defined functional state, with links to a constituent list and a transition graph (if applicable) to indicate all the transitions that lead to the state. A protein state is defined by the principal proteins interactions with other protein partners, covalent modifications on all protein components, association with small molecule ligands and cellular location. The ‘Transitions’ section shows a list of the defined transitions, with a link to detailed information on each transition—with initial and final state information, the change that occurred in the transition, process information, other comments and citations (Figure 1). A transition is defined as a biological process that causes the conversion of a protein from one state to another. The ‘Network Map’ gives a graphical representation of all the states, and the transitions between them, defined by the author for A000001 (Figure 2). The ‘Functions’ section shows that A000001 acts as an enzyme, catalyzing the conversion of Mg-ATP to cyclic AMP and pyrophosphate. Each state that catalyzes the reaction is listed, with a link to the detailed state information, and a link to detailed function information with reaction information, comments and citations (Figure 3). The ‘Protein Classes’ section shows classes defined by the author to aid in data entry and display—a class is defined as a group of three or more proteins that behave identically in a particular state.
Figure 1.
Example state transition for Adenylyl cyclase type 5 (A000001).
Figure 2.
Network map for Adenylyl cyclase type 5 (A000001).
Figure 3.
Example function for Adenylyl cyclase type 5 (A000001).
Automated data
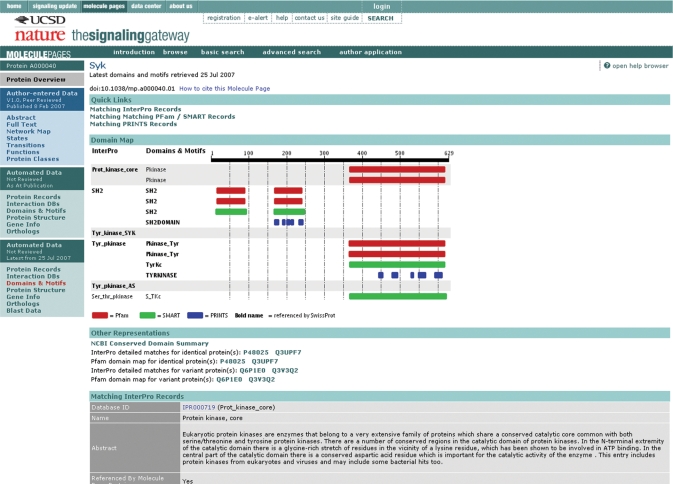
The ‘Protein Records’ section displays all the sequence database records related to particular Molecule Page. A specific record, defined by an NCBI protein GI number (3), is assigned to the Molecule Page as a base sequence. All the other sequence records listed in the same Entrez Gene (4) record are displayed, as well as any UniProt (5) and Ensembl (6) records that refer to those sequence records. The records are grouped by their specific sequence. The ‘Gene Info’ page displays pertinent information related to the Molecule from the Entrez Gene record, including any related Ensembl gene records or the sequence records within it. The ‘Domains & Motifs’ (Figure 4) section contains domain information Pfam (7) and Smart (8), pattern/motif information from PRINTS (9) and InterPRO (10) records related to the Molecule Page sequence. These records are produced using a combination of database record references, computational schemes [hmmpfam (11) and FingerPRINTScan (12)]. Matching sequence regions are given for the computational matches. The ‘Interactions’ page displays matching interaction database records from the BIND database and from the Entrez Gene database, including BIND (13), BioGRID (14) and HPRD (15) interaction records. Interactions involving likely orthologs of the Molecule Page base sequence are displayed, to provide additional information. The ‘Orthologs’ section shows genes in other select organisms that are likely orthologs of the base gene. This list is constructed using a combination of a species-specific Blast against the NCBI protein database, and database analysis of HomoloGene (16) and Ensembl homology databases. The ‘Blast Data’ section contains a list of the top Blast hits against the entire NCBI protein database. The Protein ‘Structure’ section displays the PDB (17) records that are related to the Molecule Page, either through a database reference to one of the related protein sequence records, or by a sequence match with Blast.
Figure 4.
Domain/Motif map for Syk (A000040).
In addition to all the hyperlinks to the relevant bioinformatic databases, the SGMP base sequence also links directly to the SDSC Biology Workbench (http://workbench.sdsc.edu). The Biology Workbench (18) enables a user to carry out seamlessly a variety of sequence analysis operations. The link is located on the ‘Protein Overview’ page.
EDITORIAL PROCESS
NPG and UCSD are assisted by a scientific Advisory Board and an Editorial Board. The Advisory Board provides high-level guidance and advice concerning the development of the Molecule Pages database. The Editorial Board helps the editorial team with several aspects of publishing Molecule Pages, such as identifying relevant authors, reviewers and adjudication during peer-review.
NPG manages a rigorous editorial process to ensure that expert-authored Molecule Pages are accurate and complete, and that structured data is recorded in a consistent manner. Authors either apply to contribute and are selected by NPG editors, or are commissioned by editors. After an initial editorial evaluation of an author's submission, the Molecule Page undergoes anonymous peer-review by two or three experts in the relevant field. Following peer-review the author may be required to revise their submission in light of reviewer and editor comments. Following revision the Molecule Page is critically assessed and a decision to publish is made. The Molecule Page is copy edited before publication and a Digital Object Identifier (DOI) assigned upon publication.
DATABASE IMPLEMENTATION
The SGMP is a multi-tier Enterprise Java web application. The database tier is an Oracle 10g database instance running on a Sun server. The middle tier contains business and web components and is deployed on an Oracle Components 4 Java (OC4J) application server that also runs on a Sun server. The client tier consists of a web browser running on the user's machine. The business components consist of data access objects that encapsulate database access for the web layer. The web layer consists of Java servlets and server pages complying with the J2EE (Enterprise Java) 1.3 specifications. The compute and database servers are located at the San Diego Supercomputer Center (SDSC). SDSC provides hardware, network and system administrative support.
In addition to the web forms that the general public uses for viewing the data, there are specialized web forms for the authors, reviewers and editors to perform their tasks. A password-protected user access system controls access to the specialized forms and the unpublished data for a given Molecule Page, but the general public is able to access any published data and all automated data without having to register for an account. Registration to the Signaling Gateway is, and always will be, free.
The automated data is calculated monthly. Local copies of the constituent databases are stored in custom, relational forms on the Oracle system, with the computational methods being run on Sun systems. The results of the automated data are stored in the database tier, along with the archived results of automated analysis at the time of publication. Tab-delimited files relating the Molecule Pages to protein sequence database records (e.g. UniProt, Refseq, Genbank) and gene database records (Entrez Gene and Ensembl) are provided via anonymous ftp.
The data is accessed via a browse function, a simple search engine and an advanced search engine. The simple search engine allows users to query the database using the Molecule Page ID, gene symbols, protein names and synonyms. The advanced search engine allows users to ask complex questions, such as ‘show me all functional states involving Molecule A and Molecule B.’ The advanced search engine uses the Lucene library (http://lucene.apache.org/) from the Apache Software Foundation (http://www.apache.org/)—an open-source Java toolset.
FUTURE DIRECTIONS
A wiki will be added to the web site, allowing for open comments on any given Molecule Page, as well as a living molecule summary that does not require the rigorous process of a published Molecule Page. Previously published Molecule Pages will be updated by the authors, and released as subsequent versions. We plan on adding exportable Molecule Page information, for published Molecule Pages, in XML form, as well as standard exchange formats such as BioPAX (http://www.biopax.org/). We will continue to add to the links provided in the automated data sections, addressing areas such as gene expression and phosphorylation.
ACKNOWLEDGEMENTS
We would like to acknowledge National Institutes of Health Grant [5 U54 GM62114-06], the National Institute of General Medical Science glue grant, which supported the AfCS. We thank Dr Alfred Gilman, the principle investigator of the AfCS project, as well as the members of the AfCS research team, many of whom were integral in the development and beta testing of the Molecule Pages. Warren Hedley, Yuhong Ning and Ilango Vadivelu contributed to the business and presentation tiers of the Molecule Page application, and Joshua Li contributed to the Oracle database tier.
Timo Hannay, the Publishing Director at Nature.com, was instrumental in the creating of the alliance with Nature Journal, and provides invaluable advice for the development and directions of the Signaling Gateway. Barbara Marte and Bernd Pulverer of Nature provide much needed advice on editorial decisions. Our editorial advisory board contains Pat Casey, Michael Berridge, Tony Hunter and Robin Irvine, and in addition we would like to acknowledge all our editorial board. The UCSD-Nature Signaling Gateway is funded by the National Institutes of Health Grant [1 R01 GM078005-01]. Funding to pay the Open Access publication charges for this article was provided by The National Institutes of Health Grant [1 R01 GM078005-01].
Conflict of interest statement. None declared.
REFERENCES
- 1.Gilman AG, Simon MI, Bourne HR, Harris BA, Long R, Ross EM, Stull JT, Taussig R, Bourne HR, et al. Overview of the alliance for cellular signaling. Nature. 2002;420:703–706. doi: 10.1038/nature01304. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Ning Y, Hedley W, Saunders B, Chen Y, Tindill N, Hannay T, Subramaniam S. The Molecule Page Database. Nature. 2002;420:716–717. doi: 10.1038/nature01307. [DOI] [PubMed] [Google Scholar]
- 3.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The UniProt Consortium. Entrez The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attwood TK, Blythe MJ, Flower DR, Gaulton A, Mabey JE, Maudling N, McGregor L, Mitchell AL, Moulton G, et al. PRINTS and PRINTS-S shed light on protein ancestry. Nucleic Acids Res. 2002;30:239–241. doi: 10.1093/nar/30.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonnhammer EL, Eddy SR, Birney E, Bateman A, Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scordis P, Flower DR, Attwood TK. FingerPRINTScan: intelligent searching of the PRINTS motif database. Bioinformatics. 1999;15:799–806. doi: 10.1093/bioinformatics/15.10.799. [DOI] [PubMed] [Google Scholar]
- 13.Bader GD, Betel D, Hogue CWV. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stark C, Breitkreutz B, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, et al. Human protein reference database – 2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edga R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramaniam S. The Biology Workbench – a seamless database and analysis environment for the biologist. Proteins. 1998;32:1–2. [PubMed] [Google Scholar]