Abstract
We developed a database called RNAJunction that contains structure and sequence information for RNA structural elements such as helical junctions, internal loops, bulges and loop–loop interactions. Our database provides a user-friendly way of searching structural elements by PDB code, structural classification, sequence, keyword or inter-helix angles. In addition, the structural data was subjected to energy minimization. This database is useful for analyzing RNA structures as well as for designing novel RNA structures on a nanoscale. The database can be accessed at: http://rnajunction.abcc.ncifcrf.gov/
BACKGROUND
Nucleic acid systems have proven to be very amenable to the design of nanostructures. While there are more published examples for DNA systems (1–4), RNA has also been used to self-assemble into various shapes like squares and triangles (4,5). The corners of the assembled RNA complexes are based on known helical junctions (4) or loop–loop interactions (6). These designed RNA structures highlight the importance of loop–loop motifs and helical junctions. A database of such structural elements would significantly speed up the design process.
Helical junctions are important for the structural and catalytic properties of RNAs. It has been shown, for example, that a four-way junction promotes the functional folded state of the hairpin ribozyme (7). Thus, characterizing and classifying RNA junctions can lead to a better understanding of the structural and functional capabilities of RNA.
Several databases containing RNA structures exist. The basic repository for experimentally determined nucleic acid structures is the Protein Data Bank (PDB) (8). Because RNA structures are only part of the content of the PDB, several databases provide additional information by annotating and classifying RNA structures derived from the PDB. SCOR is a structural database that contains a classification of internal and hairpin loops (9–11). Its classification scheme is based on a directed acyclic graph, allowing a node to have multiple parents. NCIR is a database of non-canonical interactions found in RNA structures (12). For each base pair type, NCIR provides information about sequence and structure contexts in which this base pair type has been found. The Nucleic Acid Database (NDB) is a database containing annotated and categorized RNA and DNA structures (13). Among other things, it provides categories for RNA junctions and DNA junctions. The provided junctions correspond to complete PDB structures and not the extracted fragments. The Metals in RNA (MeRNA) database catalogs RNA structures that are bound to metal ions (14).
Lescoute and Westhof (15) analyzed RNA structures with respect to three-way junctions. They found that three-way junctions that contain two helices that are coaxially stacked can be classified into three main families depending on the relative lengths of the connecting loop regions. Lilley (16) reviewed helical junctions of DNA and RNA. This study showed a bias towards coaxial stacking and the importance of ion interactions. Lilley et al. (17) describe a NC-IUBMB recommended nomenclature for nucleic acids junctions.
We have developed a database called RNAJunction that provides information about RNA junctions, kissing loops, internal loops and bulges in an extracted, annotated and searchable form. Our RNAJunction database is very useful for analyzing and understanding the principles of RNA structure formation. It is to our knowledge the only currently available database that also contains extracted RNA kissing loop elements. Its unique search capability allows the user to identify RNA junctions based on (among other criteria) inter-helical angles, which makes it an important resource for the design of novel RNA nanostructures from building blocks.
CONSTRUCTION AND CONTENT
Junction scanning algorithm
Our definition of an n-way RNA (or DNA) junction is best explained with the help of Figure 1, a schematic view of a hypothetical three-way junction. One can see from that example that an extracted n-way junction consists of n strands and n connector helices. Note that the original structure from which the junction is extracted might consist of only one strand. Also, internal loops and bulges can be viewed as special cases of ‘two-way’ junctions. Our definition of RNA helix junctions is geared towards the design of RNA nanostructures. From this standpoint, an important property of a junction is the relative orientation of its ‘connector’ helices.
Figure 1.
Schematic drawing of a three-way junction.
We developed a Java program, ‘JunctionScanner,’ for detecting, extracting and analyzing RNA junctions, kissing loops, internal loops and bulges from PDB coordinate files. The algorithm uses the RNAView (18) base pairing patterns to detect groups of interconnected helices corresponding to RNA junctions from a given structure in PDB format. Non-canonical base pairs are allowed. Given the list of base pairs, a list of all bulge-free helices is generated (a helix containing a bulge is represented as two helices). Each strand of each helix is used as a starting point of a ‘path’ as follows: using a start strand and helix position, the strand is followed in the 5′ to 3′ direction to the next downstream helix. The opposing strand from this subsequent helix is now followed from its 5′ to its 3′ end to yet another helix and so forth. If one arrives in this fashion back at the helix one started with (a circular path), one can conclude that the helices of the path form a junction (compare Figure 1). Note that hairpin loops are currently not considered. The JunctionScanner program extracts the coordinate data corresponding to each unique detected junction. Also, the local orientation of each helix is approximated by superposing a helix represented by the idealized positions of C4′ atoms (19). This permits the computation of the angle between the direction-vectors of any two helices.
Several filters are applied to the initial set of solutions. We found that the number of extracted structural elements depends on the cutoff-parameters used by the various filters. Depending on the use, one wants to apply strict or relaxed filter parameters. To account for different applications, we generated three different sets of extracted structural elements using parameter sets ranging from ‘least strict’ to ‘most strict’ (the values of the different parameter sets are shown in Table 1). First, the structural elements are not allowed to contain non-standard bases. The connector helices have to consist of at least n base pairs (n being two, three or four, Table 1). An idealized helix has to be fitted onto the connector helices with an RMSD being smaller than the chosen cut-off of 2.5 or 3.0 Å (Table 1). The sum of the number of nucleotides of the junction loop regions may not exceed 50 nt. Another optional filter is the ‘corridor-filter’: corridors are defined as cylindrical regions (3 Å radius) with their base being located at the center of the junction's helix ends. The cylinders are oriented such that they point outward in the direction of the respective helix (the geometry involved in the corridor filter is shown schematically in Figure S2 in the Supplementary Data). The corridor regions are checked for steric clashes with atoms of the junction. This steric clash check limits the retrieved junctions to cases that are potentially useful for the ‘mosaic unit’ approach of building larger structures from building blocks (20–22).
Table 1.
Cut-off values for different filtering parameter sets called ‘most strict’, ‘medium’ and ‘least strict’
| Most strict | Medium | Least strict | |
|---|---|---|---|
| Minimum helix length (bp) | 4 | 3 | 2 |
| RMS for helix fit (Å) | 2.5 | 3.0 | 3.0 |
| ‘Corridor’ filter | Yes | Yes | No |
Note that the structural elements corresponding to the most strict parameter set are required to possess helices of lengths four (or greater) in the original structure, however the corresponding extracted coordinate files contain only three base pairs.
Kissing loop detection algorithm
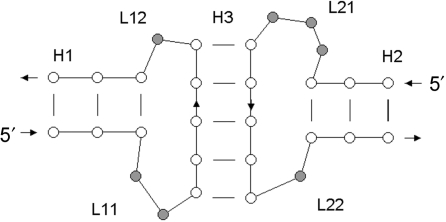
The detection of kissing loops is handled in a manner that is similar to junction detection. Again, in a first step a set of all RNA double helices is generated. Each set of three helices that interact according to the connectivity shown in Figure 2 is considered a kissing loop interaction. Using the schematic drawing of Figure 2 as an example, the loop region of helix H1 consists in our definition of the loops L11, L12 as well as one strand of helix H3. For each pair of stems a and b, the algorithm tries to find a third helix c, that is located in the loop region of stems a and b. To reduce the computational cost, we applied further constraints by requiring that the helix ends of helices a and b must be closer than 50 Å. The other filters used for junctions are also applied.
Figure 2.
Schematic drawing of a kissing loop. Each circle represents one nucleotide; the single-stranded regions are indicated in gray.
Data preparation
We downloaded 1176 structures from the PDB database that contain RNA (as of June 2007). The base pairing patterns were ascertained using RNAView (18). The JunctionScanner program described in the previous section was used to parse the PDB and RNAView data. For each identified structural element the JunctionScanner program generates two files: a file containing the extracted element in PDB format, and a text file describing all identified properties (such as helix orientation, inter-helix angles, nomenclature, nucleotide sequences, residue indices; the full list of properties is shown in the Supplementary Data). The JunctionScanner algorithm was applied five times to each coordinate input file to avoid the unlikely chance that a helix was missed due to the random component inherent in the helix-fitting algorithm. The algorithm identified 258 kissing loop structures, 9357 internal loops and bulges, 2065 three-way junctions, 1091 four-way junctions, 462 five-way junctions, 70 six-way junctions, 23 seven-way junctions, one eight-way junction and one nine-way junction. Five hundred and fifty-nine different PDB files contained structural elements meeting the minimum filter criteria described earlier. The counts for the number of identified junctions depend on how strict the parameters for the junction-detection algorithm are chosen. The counts mentioned above correspond to the ‘least strict’ parameter set. Counts for the different junction types and junction scanner parameter sets are listed in Table 2. If a structural element is identified using several different filter parameter sets, only the version corresponding to the strictest parameter set is stored in the database, thus avoiding unnecessary redundancy.
Table 2.
Counts of identified RNA junctions, internal loops, bulges and kissing loops The counts are given for three different filter parameters
| Structural element | Most strict | Medium strict | Least strict | Reduced redundancy |
|---|---|---|---|---|
| Kissing loop | 98 | 206 | 258 | 51 |
| Internal loop/bulge | 3133 | 5635 | 9357 | 1837 |
| 3-way junction | 731 | 1412 | 2065 | 431 |
| 4-way junction | 280 | 490 | 1091 | 272 |
| 5-way junction | 109 | 237 | 462 | 97 |
| 6-way junction | 4 | 6 | 70 | 17 |
| 7-way junction | 45 | 55 | 61 | 16 |
| 8-way junction | 0 | 1 | 1 | 1 |
| 9-way junction | 0 | 0 | 1 | 1 |
The last column shows the counts for the reduced-redundancy least-restricted data set whose representative structures can be part of either the most-strict, medium-strict or least-strict data set.
A simple scheme for optionally working with a reduced redundancy data set was adopted. All structural elements of the same type, consisting of the same sequences were clustered using single linkage clustering and a structural superposition cutoff of 3.0 Å DRMS (C4′ atom positions). For each cluster, one of its members is chosen to be the representative structure. The user can choose to search the original data or only the representative structures. This reduces the number of structural elements by a factor of about four (compare Table 2). Analyzing the structural variation within clusters, we found that in all but three cases the DRMS of the cluster members with respect to their cluster representative is smaller then 3.0 Å.
Web and database tier
The results of the JunctionScanner applied to PDB coordinate files was entered into a MySql relational database. These data include the quantitative analysis, sequence, geometric conformation and citation for the identified structural elements as well as references to the parent structure from the PDB and the extracted coordinate data (the full list of stored properties is given in the Supplementary Data). The web presentation tier is implemented using PHP. Images of the secondary structure (generated using RNAView) and tertiary structure [generated using Raster3D (23)] are available. The 3D coordinates of the structural element can be viewed interactively using the JMol Java applet. The text output of the JunctionScanner run is also provided and contains more detailed information such as the local coordinate systems of the fitted idealized helices. Additional data is included in the form of structures obtained by molecular mechanics minimization, calculated using Amber 8.0 (24) and the Cornell force field ff99 (25). Analyzing the minimized structures, we found that 68% of them have an RMSD of <1 Å compared to their respective original structure; 31% have an RMSD between 1 and 2 Å, 0.7% exhibit an RMSD between 2 and 3 Å, 0.0003% show an RMSD between 3 and 5 Å. We plan to supplement the RNAJunction database also with molecular dynamics simulation results in the future.
The essential tables underlying the RNAJunction database correspond to junctions, helices, strands, references and angles. In this way, helices and junctions, for example, are related through a many-to-one relationship, allowing in principle for junctions of arbitrarily large order. More detailed information about the design of the database tables is provided in the Supplementary Data. Internal loops, bulges and kissing loops are internally considered a special case of ‘two-way’ junctions; a flag indicates the different strand connectivity of a kissing loop compared to an internal loop.
UTILITY
The aim of this database is 2-fold: first, it is an important resource for RNA nanodesign, which requires a library of known RNA structural elements for the development of novel RNA structures (20,26). Second, we envision the database to be useful for analysis, in particular for understanding the principles of RNA structure and folding.
RNAJunction (http://rnajunction.abcc.ncifcrf.gov/) is publicly available and its data can be accessed through a user-friendly website. The RNAJunction database contains detailed information about RNA structural elements (currently junctions, internal loops, bulges and kissing loops). Using the web interface, the user can search for structural elements based on their NC-IUBMB nomenclature (17), type of junction, inter-helical angles, PDB identifier, the RNAJunction identifier, sequence, experimental method, author of the corresponding published structure or parts of the text contained in the header records of the PDB structure. The results of a search are displayed by listing a summary of all matching structural elements. Users may then click on a database identifier which points to a page providing more detailed information about a particular structural element.
The database provides, in its detailed view, information about the sequences of a structural element, its NC-IUBMB nomenclature, inter-helix angles. Even more detailed information, such as the orientation of the connecting helices can be obtained by following the ‘JunctionScanner output’ link. Web links are provided for a literature citation as well as various related databases, such as the PDB (27), SCOR (9,10) and MMDB (28). Using the JMol Java applet (http://www.jmol.org/), the user can interactively display a 3D representation of a junction or kissing loop. As an example of the utility of the database, we show the result page of a kissing loop structure (PDB id 2BJ2, Figure 3). This structural element has been used for the design of an RNA hexagonal ring (6). The RNAJunction database annotates this structure to possess an inter-helix angle of 122° (see Figure 3 bottom). Thus, the added information makes it immediately clear that this structural element is a good candidate for designing a hexagonal structure requiring angles of ∼120°.
Figure 3.
Result page of an RNA loop–loop interaction derived from PDB structure 2BJ2.
AVAILABILITY AND REQUIREMENTS
The RNAjunction database is freely available at http://rnajunction.abcc.ncifcrf.gov
General queries can be performed using virtually any web browser; displaying 3D structures with JMol requires the browser to be able to run Java applets. The database is available for download (structures, JunctionScanner output, images) upon request. Periodic updates will be made based upon the availability of new RNA structures.
ACKNOWLEDGEMENTS
We thank Luc Jaeger and Christine Viets for valuable suggestions and Mary O’Connor for help with the web interface. We wish to thank the Advanced Biomedical Computing Center (ABCC) at the NCI for their computing support and for hosting the web and database servers. This publication has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. N01-CO-12400. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Funding to pay the Open Access publication charges for the article was provided by National Cancer Institute (NCI).
Conflict of interest statement. None declared.
REFERENCES
- 1.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 2.Sa-Ardyen P, Jonoska N, Seeman NC. Self-assembly of irregular graphs whose edges are DNA helix axes. J. Am. Chem. Soc. 2004;126:6648–6657. doi: 10.1021/ja049953d. [DOI] [PubMed] [Google Scholar]
- 3.Chen JH, Seeman NC. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 4.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 5.Guo S, Tschammer N, Mohammed S, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum. Gene. Ther. 2005;16:1097–1109. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yingling YG, Shapiro BA. Computational design of an RNA hexagonal nanoring and an RNA nanotube. Nano Lett. 2007;7:2328–2334. doi: 10.1021/nl070984r. [DOI] [PubMed] [Google Scholar]
- 7.Wilson TJ, Nahas M, Ha T, Lilley DM. Folding and catalysis of the hairpin ribozyme. Biochem. Soc. Trans. 2005;33:461–465. doi: 10.1042/BST0330461. [DOI] [PubMed] [Google Scholar]
- 8.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klosterman PS, Hendrix DK, Tamura M, Holbrook SR, Brenner SE. Three-dimensional motifs from the SCOR, structural classification of RNA database: extruded strands, base triples, tetraloops and U-turns. Nucleic Acids Res. 2004;32:2342–2352. doi: 10.1093/nar/gkh537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klosterman PS, Tamura M, Holbrook SR, Brenner SE. SCOR: a structural classification of RNA database. Nucleic Acids Res. 2002;30:392–394. doi: 10.1093/nar/30.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura M, Hendrix DK, Klosterman PS, Schimmelman NR, Brenner SE, Holbrook SR. SCOR: structural classification of RNA, version 2.0. Nucleic Acids Res. 2004;32:D182–D184. doi: 10.1093/nar/gkh080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagaswamy U, Larios-Sanz M, Hury J, Collins S, Zhang Z, Zhao Q, Fox GE. NCIR: a database of non-canonical interactions in known RNA structures. Nucleic Acids Res. 2002;30:395–397. doi: 10.1093/nar/30.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh SH, Srinivasan AR, Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stefan LR, Zhang R, Levitan AG, Hendrix DK, Brenner SE, Holbrook SR. MeRNA: a database of metal ion binding sites in RNA structures. Nucleic Acids Res. 2006;34:D131–D134. doi: 10.1093/nar/gkj058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lilley DM. Structures of helical junctions in nucleic acids. Q. Rev. Biophys. 2000;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 17.Lilley DM, Clegg RM, Diekmann S, Seeman NC, Von Kitzing E, Hagerman PJ. A nomenclature of junctions and branchpoints in nucleic acids. Nucleic Acids Res. 1995;23:3363–3364. doi: 10.1093/nar/23.17.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H, Jossinet F, Leontis N, Chen L, Westbrook J, Berman H, Westhof E. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 2003;31:3450–3460. doi: 10.1093/nar/gkg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aalberts DP, Hodas NO. Asymmetry in RNA pseudoknots: observation and theory. Nucleic Acids Res. 2005;33:2210–2214. doi: 10.1093/nar/gki508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westhof E, Masquida B, Jaeger L. RNA tectonics: towards RNA design. Fold Des. 1996;1:R78–R88. doi: 10.1016/S1359-0278(96)00037-5. [DOI] [PubMed] [Google Scholar]
- 21.Tsai CJ, Zheng J, Aleman C, Nussinov R. Structure by design: from single proteins and their building blocks to nanostructures. Trends Biotechnol. 2006;24:449–454. doi: 10.1016/j.tibtech.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Zheng J, Zanuy D, Haspel N, Tsai CJ, Aleman C, Nussinov R. Nanostructure design using protein building blocks enhanced by conformationally constrained synthetic residues. Biochemistry. 2007;46:1205–1218. doi: 10.1021/bi061674a. [DOI] [PubMed] [Google Scholar]
- 23.Merritt EA, Murphy ME. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 24.Case DA, Cheatham T.E., III, Darden T, Gohlke H, Luo R, Merz K.M., Jr, Onufriev A, Simmerling C, Wang B, et al. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 26.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouranov A, Xie L, de la Cruz J, Chen L, Westbrook J, Bourne PE, Berman HM. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34:D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Anderson JB, Chen J, Geer LY, He S, Hurwitz DI, Liebert CA, Madej T, Marchler G, et al. MMDB: Entrez's 3D-structure database. Nucleic Acids Res. 2002;30:249–252. doi: 10.1093/nar/30.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]