Abstract
The Yeast search for transcriptional regulators and consensus tracking (YEASTRACT) information system (www.yeastract.com) was developed to support the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Last updated in September 2007, this database contains over 30 990 regulatory associations between Transcription Factors (TFs) and target genes and includes 284 specific DNA binding sites for 108 characterized TFs. Computational tools are also provided to facilitate the exploitation of the gathered data when solving a number of biological questions, in particular the ones that involve the analysis of global gene expression results. In this new release, YEASTRACT includes DISCOVERER, a set of computational tools that can be used to identify complex motifs over-represented in the promoter regions of co-regulated genes. The motifs identified are then clustered in families, represented by a position weight matrix and are automatically compared with the known transcription factor binding sites described in YEASTRACT. Additionally, in this new release, it is possible to generate graphic depictions of transcriptional regulatory networks for documented or potential regulatory associations between TFs and target genes. The visual display of these networks of interactions is instrumental in functional studies. Tutorials are available on the system to exemplify the use of all the available tools.
OVERVIEW
YEASTRACT (Yeast Search for Transcriptional Regulators And Consensus Tracking; www.yeastract.com) was originally proposed (1) to make publicly available up-to-date information on documented regulatory associations between TFs and target genes, as well as between TFs and DNA binding sites, in Saccharomyces cerevisiae. Additionally, it provides a set of bioinformatics tools that facilitate the full exploitation of the data. Although part of the data was obtained from existing yeast data repository like the S. cerevisiae genome database (SGD) (2), the gene ontology (GO) consortium (3) and the regulatory sequences analysis tools (RSAT) (4), all the data on gene regulation was gathered based on exhaustive literature analysis.
The value of YEASTRACT comes from the integration of complete and up-to-date regulatory information, with a number of analysis methods and computational tools. The usefulness of YEASTRACT for the analysis of gene lists, in particular those coming from gene expression analysis by microarrays, also distinguishes this information system from others. Although other databases have also made available information about regulatory mechanisms in yeast and other organisms [e.g. MYBS (5) and TRANSFAC (6)] or computational tools for the analysis of promoter regions [RSAT (4)], YEASTRACT is the system that most seamlessly integrates extensive regulation data and computational tools for the analysis of this information.
The database presently contains more than 30 990 regulatory associations between genes and TFs, based on more than 1000 bibliographic references. These include five papers describing global ChIP analysis (7–11), which document 75% of the gathered regulatory associations and one microarray analysis on the effect of the deletion of 55 TFs (12), documenting 15% of these regulatory associations. The results of hundreds of other articles describing more detailed molecular analysis and revealing many regulatory associations that were not detected by global experiments are also included in this version of the system. The explosion of the scientific knowledge in the field of transcriptional regulation led to a 300% increase on the actual number of regulatory associations in the system, with respect to the first release. Each regulation has been annotated manually, after expert examination of the relevant references. The database presently contains 284 specific DNA binding sites for 108 characterized TFs. The total number of TFs in the database is 170, which corresponds to all genes that are identified as TFs at SGD.
A comprehensive description of the content and structure of YEASTRACT has been presented in the first publication of this system (1). At a high level, the internal structure of the database is organized around the concept of gene, protein and binding site (consensus) and these three concepts are related by regulation relations. These relations document the associations between TFs and target genes and can be of two types: documented and potential. In the first release, the system made available a set of queries to facilitate the exploitation of the gathered data when solving a number of biological questions, in particular those that involve the analysis of global gene expression results. In the first 6 months of 2007, researchers from more than 300 different institutions, from 70 different countries, have performed over 90 000 queries using YEASTRACT. The number of queries in this period has already reached the total number of queries performed during 2006.
In this new release, the available queries and additional utilities were reorganized to simplify their use, maintaining the user-friendly interface and functionality, which were already present in the original release of the system. YEASTRACT has already demonstrated its usefulness as a tool to support research on transcription regulation processes in yeast (13). Nonetheless, this release significantly extends the capabilities of the system by connecting it with a number of data processing tools that will significantly increase its usefulness. YEASTRACT now includes DISCOVERER, a system that enables the user to search for common motifs in the promoter region of genes, using efficient algorithms for structured motif discovery and to automatically compare the results with the transcription factor binding sites (TFBS) described in YEASTRACT. Pattern matching algorithms were also included to enable the user to search the promoter region of one or more genes, for one or more DNA motifs, specified using a number of different representations.
Another important new feature is the possibility to identify and display transcription regulatory networks (TRNs) for documented and potential regulatory associations between TFs and target genes. This feature supports the analysis of regulatory mechanisms, based on permanently up-to-date, manually checked, information. Such an analysis will, in the future, support mechanisms for the inference of TRNs in S. cerevisiae, one of the main strategic objectives of this project.
DISCOVERER
The precise coordinated control of gene expression is accomplished by the interplay of multiple regulatory mechanisms. The transcriptional machinery is recruited to the promoter leading to the transcription of the downstream gene through the binding of transcription regulatory proteins to short nucleotide sequences occurring in gene promoter regions. To support the analysis of the promoter sequences in the yeast genome, a set of software tools is available in DISCOVERER. DISCOVERER provides tools for motif extraction, which consists on the identification of de novo binding site consensus sequences from a given set of non-coding DNA sequences (such as the promoter regions of a gene). DISCOVERER contains two distinct structured motif discovery algorithms: MUSA (14) and RISO (15).
When the algorithms finish, the user receives, by e-mail, a link to a web page (Figure 1) where it is possible to download the complete list of motifs found, ordered by their P-value and showing the proportion of sequences containing each motif (the quorum). The motifs identified are also clustered in families, represented by a position weight matrix (PWM) description. This assembling of individual motifs into families of motifs is very useful in reducing the number of motifs, leading to a more tractable output and to a more intuitive motif representation. A new algorithm for the motif assembling problem was developed (16), since this is a very difficult problem in its own right (17,18). From the output page it is also possible to download the list of motifs and the PWM description for each family.
Figure 1.
Sample pages showing the motif finder output presenting a PWM for each motif family found and the match output obtained by querying the database for TFs binding sites that match the selected family. These results were obtained for the sample data available in the algorithm's input page. A detailed analysis of these results is available in the DISCOVERER tutorial.
Each PWM can be selected, to be compared with the TFBS that are described in the YEASTRACT database. The input PWM is locally aligned [using the Smith–Waterman local alignment algorithm (19)] with each of the TFBS PWM, using a specific column distance metric from a set of options available. The list of the top twenty scoring alignments is displayed for user inspection.
NEW REFINEMENTS
Pattern matching
YEASTRACT now makes available pattern matching methods, supporting the search for one or more nucleotide sequences (e.g. TFBS) within the promoter region of chosen genes, thus leading to the identification of putative target genes for specificTFs. However, the TFBS, used for pattern matching, have to be provided by the user. The query string may be a simple nucleotide sequence, a sequence containing IUPAC nucleotide code or even a sequence containing regular expression elements. The forward and reverse strands of the promoters are searched for the input motifs.
This search returns a list of genes in whose promoters the patterns were found, including the number of occurrences in each promoter. The patterns that matched the promoter sequences and their locations in the promoters are also displayed.
The queries that were refined, or that are new, using the extended pattern matching capability, are the following ones:
Search by DNA motif
Find TFBs
Search Motifs on Motifs
Transcriptional regulatory networks
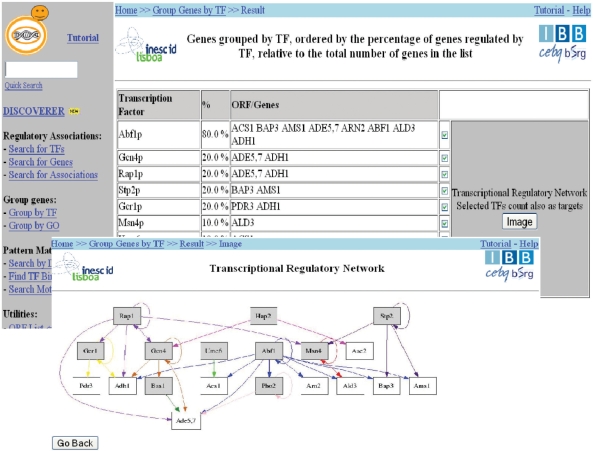
Recent studies in data collection and analysis (7,20,21) have shown that the information needed to understand regulatory networks must come from the integration of different sources, such as genomic sequence data, genome-wide transcription data, structural information and biological literature. The comprehensive data on regulatory associations available in YEASTRACT makes it possible to identify and visualize TRNs for documented (i.e. described in the literature) and potential (a known binding site is present in the promoter region) regulatory associations between TF and target genes (Figure 2).
Figure 2.
Sample pages showing the regulation graph obtained from the ‘Group by TF’ query. Shaded squares represent TF and white squares represent genes. Arrows represent interactions between each TF and target genes.
These transcriptional regulatory networks and, in particular, the documented ones, correspond to static regulatory networks in S. cerevisiae, since the evidence for the regulatory associations has been described for different processes and experimental conditions.
The generation of TRNs through the queries ‘Group by TF’ and ‘Generate Regulation Matrix’, enables the analysis of regulatory mechanisms, supported by up-to-date, manually checked, information. Such an analysis will, in the future, support the inference of mechanisms underlying gene regulatory networks in S. cerevisiae.
ACKNOWLEDGEMENTS
The information about yeast genes other than documented regulations, potential regulations and the TFBS contained in YEASTRACT was gathered from SGD, the GO Consortium and RSAT. We are also grateful to colleagues and friends from the yeast community for their encouragement and suggestions. This work was supported by FEDER, FCT and the POSI, POCTI and PDCT programs (projects POSI/EIA/57398/2004, POCTI/BIO/56838/2004 and PDTC/BIO/72063/2006, and PhD or post-doctoral grants—SFRH/BD/32965/2006, SFRH/BD/29246/2006, BPD/28625/2006, BPD/5649/01, SFRH/BD/17456/2004, SFRH/BD/23437/2005—to PM, NDM, MCT, ST, NM, AL, respectively). Funding to pay the Open Access publication charges for this article was provided by Fundação para a Ciência e a Tecnologia, project DBYEAST (POSI/EIA/57398/2004).
Conflict of interest statement. None declared.
REFERENCES
- 1.Teixeira MC, Monteiro P, Jain P, Tenreiro S, Fernandes AR, Mira NP, Alenquer M, Freitas AT, Oliveira AL, et al. The YEASTRACT database: a tool for the analysis of transcriptional relatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34(Database Issue):D446–D451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong EL, Balakrishnan R, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Livstone M, et al. Saccharomyces Genome Database. 2007. ftp://ftp.yeastgenome.org/yeast/
- 3.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai HK, Chou MY, Shih CH, Huang GTW, Chang TH, Li WH. MYBS: a comprehensive web server for mining transcription factor binding sites in yeast. Nucleic Acids Res. 2007;35(Web Server Issue):W221–W226. doi: 10.1093/nar/gkm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, Krull M, Matys V, Michael H, et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 8.Horak CE, Luscombe NM, Qian J, Bertone P, Piccirrillo S, Gerstein M, Snyder M. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 2002;16:3017–3033. doi: 10.1101/gad.1039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Workman CT, Mak HC, McCuine S, Tagne JB, Agarwal M, Ozier O, Begley TJ, Samson LD, Ideker T. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borneman AR, Zhang ZD, Rozowsky J, Seringhaus MR, Gerstein M, Snyder M. Transcription factor binding site identification in yeast: a comparison of high-density oligonucleotide and PCR-based microarray platforms. Funct. Integr. Genomics. 2007;7:335–345. doi: 10.1007/s10142-007-0054-7. [DOI] [PubMed] [Google Scholar]
- 12.Chua G, Morris QD, Sopko R, Robinson MD, Ryan O, Chan ET, Frey BJ, Andrews BJ, Boone C, et al. Identifying transcription factor functions and targets by phenotypic activation. Proc. Natll Acad. Sci. USA. 2006;103:12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teixeira MC, Fernandes AR, Mira NP, Becker JD, Sá-Correia I. Early transcriptional response of Saccharomyces cerevisiae to stress imposed by the herbicide 2,4-dichlorophenoxyacetic acid. FEMS Yeast Res. 2006;6:230–248. doi: 10.1111/j.1567-1364.2006.00041.x. [DOI] [PubMed] [Google Scholar]
- 14.Mendes ND, Casimiro AC, Santos PM, Sá-Correia I, Oliveira AL, Freitas AT. MUSA: a parameter free algorithm for the identification of biologically significant motifs. Bioinformatics. 2006;22:2996–3002. doi: 10.1093/bioinformatics/btl537. [DOI] [PubMed] [Google Scholar]
- 15.Carvalho AM, Freitas AT, Oliveira AL, Sagot M-F. An efficient algorithm for the identification of structured motifs in DNA promoter sequences. IEEE/ACM Trans. Comput. Biol. Bioinform. 2006;3:126–140. doi: 10.1109/TCBB.2006.16. [DOI] [PubMed] [Google Scholar]
- 16.Francisco AP, Freitas AT, Oliveira A. 2007. Finding motif communities. INESC-ID Technical report 15/2007. [Google Scholar]
- 17.Mahony S, Auron P, Benos PV. DNA familial binding profiles made easy: comparison of various motif alignment and clustering strategies. PLoS Comput. Biol. 2007;3:578–591. doi: 10.1371/journal.pcbi.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kankainen M, Pehkonen P, Rosenstom P, Toronen P, Wong G, Holm L. POXO: a web-enabled tool series to discover transcription factor binding sites. Nucleic Acids Res. 2006;34(Web Server issue):W534. doi: 10.1093/nar/gkl296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 20.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 21.Yeger-Lotem E, Sattath S, Kashtan N, Itzkovitz S, Milo R, Pinter RY, Alon U, Margalit H. Network motifs in integrated cellular networks of transcription-regulation and protein-protein interaction. Proc. Natl Acad. Sci. 2004;101:5934–5939. doi: 10.1073/pnas.0306752101. [DOI] [PMC free article] [PubMed] [Google Scholar]