Abstract
The PlantTribes database (http://fgp.huck.psu.edu/tribe.html) is a plant gene family database based on the inferred proteomes of five sequenced plant species: Arabidopsis thaliana, Carica papaya, Medicago truncatula, Oryza sativa and Populus trichocarpa. We used the graph-based clustering algorithm MCL [Van Dongen (Technical Report INS-R0010 2000) and Enright et al. (Nucleic Acids Res. 2002; 30: 1575–1584)] to classify all of these species’ protein-coding genes into putative gene families, called tribes, using three clustering stringencies (low, medium and high). For all tribes, we have generated protein and DNA alignments and maximum-likelihood phylogenetic trees. A parallel database of microarray experimental results is linked to the genes, which lets researchers identify groups of related genes and their expression patterns. Unified nomenclatures were developed, and tribes can be related to traditional gene families and conserved domain identifiers. SuperTribes, constructed through a second iteration of MCL clustering, connect distant, but potentially related gene clusters. The global classification of nearly 200 000 plant proteins was used as a scaffold for sorting ∼4 million additional cDNA sequences from over 200 plant species. All data and analyses are accessible through a flexible interface allowing users to explore the classification, to place query sequences within the classification, and to download results for further study.
INTRODUCTION
A common goal of current plant genomics research is to establish an expandable platform for global classification and analysis of plant gene family space. A large fraction of genes in plant genomes are the product of duplication and novel gene creation processes that have occurred within plants over their 500-million-year history. Gene classifications that attempt to capture all of eukaryote diversity typically provide a poor representation of plant gene sets. With more than a dozen plant genomes scheduled for completion over the next two years, and many additional genome and transcriptome projects being initiated, there is a need for flexible, gene family-focused databases that provide rich toolsets for comparative analyses of plant genomes. Comparative analyses of the modeled proteomes for sequenced genomes can help verify gene content and elucidate the process of gene duplication and functional diversification. Cross-validation of gene models for available plant genomes and nucleotide sequence translations of EST sets for other plant species can be achieved through clustering and similarity analyses involving whole-genome sequences and large EST sets [e.g. (3–5); TIGR Plant Transcript Assemblies, (6)].
The PlantTribes database is a global classification of genes from all of the five sequenced plant genomes: Arabidopsis thaliana, Carica papaya (papaya), Medicago truncatula (barrel medic, 60% sequenced), Populus trichocarpa (poplar) and Oryza sativa (rice). The database also contains unigene sets from the TIGR Plant Transcript Assemblies (6), which includes ∼4 million sequences from more than 200 species, that facilitates a wide range of comparative study of plant genes and gene families. PlantTribes offers a unique view of objectively defined gene families that facilitates comparative analyses of plant genomes. For example, our database allows one to identify all gene families of a given size in a species and quickly assess the range of copy numbers for closely related genes in other plant genomes. Families that have remained stable in size, or have proliferated greatly in one genome compared to another can easily be identified. In our own research, this type of analysis has aided interpretation of gene family stability and diversification in the face of gene and whole genome duplications (e.g. 24, 25, 30, 31). Integration of expression data, linked seamlessly to the tribe gene classification, will facilitate studies of expression divergence following gene duplication (e.g. 17). PlantTribes can aid comparative analyses by serving as a scaffold of gene families into which users can sort their genes of interest. We have devised search and query tools that allow users to access this information, making it possible to investigate the evolution of plant genomes through analysis of the scaffold itself and sequences sorted into the scaffold.
DATABASE PRODUCTION
Sequences were downloaded from each of the five sequenced angiosperm species including 31 921 gene models from A. thaliana (TAIR, version 7.0), 25 536 from C. papaya (version 1.0, complete), 40 567 from M. trunculata (IMGA, version 1.0, 60% complete), 45 555 from P. trichocarpa (JGI, version 1.0) and 66 710 from O. sativa (TIGR, version 5.0). The Carica and Medicago genome-sequencing projects are underway; the data for these species were included with the protein scaffold and results for these species will go ‘live’ for public access following the publication of these genomes. As summarized in Figure 1, we compared the predicted proteins for all five species in an all-against-all BLASTP (e = 1e − 10, b = 10 000) using the NCBI BLAST package (7). MCL clustering was then performed at low, medium and high stringencies (Inflation, I = 1.2, 3.0, 5.0, respectively) to produce the sets of objectively defined gene families (tribes).
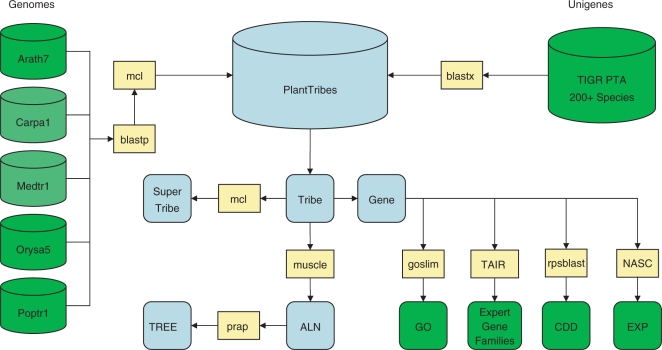
Figure 1.
PlantTribes database production. Schematic diagram detailing the process of creating the PlantTribes database. External datasets are indicated in green, ‘results’ in blue, and software in yellow. First, an all-against-all BLASTP of five sequenced plant genomes is conducted with the results sent to MCL. Taxon abbreviations: Arath7 (Arabidopsis thaliana), Carpa (Carica papaya), Medtr1 (Medicago truncatula, currently 60% complete), Orysa5 (Oryza sativa) and Poptr1 (Populus trichocarpa). Darker green for Carica and Medicago indicate that although these genomes were included in the genome scaffold, tribe results for these species will not be accessible through the web interface of PlantTribes until the public release of these genomes. Tribes are produced at low, medium and high stringencies and are annotated using Gene Ontology (GO), NCBI Conserved Domain Database (CDD) and expression data from NASCArrays (EXP). A second round of MCL clustering is performed on all tribes to group related tribes, called SuperTribes. For all tribes, protein and DNA alignments and maximum-likelihood phylogenetic trees using prap are generated. Unigene sets from the TIGR Plant Transcriptome Assemblies are searched against the fully sequenced genomes and are automatically sorted into respective tribes.
A second iteration of MCL was conducted in order to connect distant, but potentially related gene clusters which we define as SuperTribes. In order to construct SuperTribes, we computed both the average and minimum e-value between all pairs of tribes and used these as the input matrix for MCL. In addition, we ran MCL with low, medium and high inflation values to generate SuperTribe clusters at the three different stringencies. In total, there are 18 SuperTribe classifications for users to access and compare (i.e. 3 original tribe stringencies × 3 super tribe stringencies × 2 metrics ∼ average/minimum e-value).
In order to annotate each tribe, we used additional information connected to all member genes according to the following criteria: gene ontology (GO), presence of domains, manually curated gene families and common word patterns associated with the gene descriptions within a tribe. We downloaded the gene_ontology.v1.2.obo, goslim_plant.obo and gene_association.TAIR flat files (8) and used the map2slim.pl script to create a GO slim database for the Arabidopsis genes in each tribe. To annotate our tribes by domain information, we downloaded NCBI's Conserved Domain Database (CDD) (9) and used the formatrpsdb (default parameters, with f = 9.82, S = 100.0) utility to index the domains. We then searched all protein sequences from the five genomes using rpsblast (default parameters, with e-value = 1e − 5). To annotate tribes according to manually curated gene families, we downloaded gene_family_tab_121906.txt from TAIR, which includes 996 gene families that include 8331 genes. Finally, a Perl script was used to extract all gene descriptions within a tribe, and determined the most common words within the tribe, keeping track of the relative position of each word, using only the top five words. Therefore, each tribe has a composite annotation defined by each of the four criteria.
The resulting constellation of gene family tribes was used as a scaffold for plant gene space onto which roughly 4 million unigene sequences were sorted. These unigenes, derived from over 11 million ESTs, were downloaded from TIGR PTA (http://plantta.tigr.org). In addition, we sorted the predicted proteomes of Chlamydomonas reinhardtii (green alga; JGI, version 3) and Physcomitrella patens (moss; JGI, version 1). We searched the five sequenced proteomes using a blastx search (e-value = 1e − 5) for the unigene sequences and a blastp (e-value = 1e − 1) search for the distantly related Physcomitrella and Chlamydomonas proteomes.
Phylogenetic analysis pipeline
A sequence alignment and phylogenetic analysis pipeline included the following steps. We generated fasta files of both amino acid and DNA sequences (CDS) for each tribe. Each amino acid file was aligned using the MUSCLE alignment program (10). We then forced the DNA sequences onto the amino acid alignments using custom Perl/Bioperl scripts.
Phylogenetic trees were built using a fast maximum-likelihood ratchet approach (Morrison, D.A. (in press) Increasing the efficiency of searches for the maximum likelihood tree in a phylogenetic analysis of up to 150 nucleotide sequences. Syst. Biol., in press) as newly implemented in PRAP (11) v.2.0 for this study. PRAP generated command files that were handed over to PAUP (12). The heuristics involves (i) rapidly getting a starting tree not too far from the optimal score; (ii) move rapidly to a (near-) optimal tree island, (iii) getting the best tree within the island. Step (i) was achieved by calculating a BioNJ tree using LogDet distances, followed by one round of NNI and then one round of SPR branch swapping, optimizing the substitution model parameters between these steps. Similar to the parsimony ratchet (13), step (ii) included alternating between branch swapping on the original matrix and branch swapping on a matrix with 25% of characters upweighted. Unlike in Nixon's strategy for parsimony, SPR branch swapping was used, only 10 iterations were performed, and during the weighted analyses, only one tree was saved. In particular for datasets with low levels of phylogenetic signal, this strategy was found to be more successful (Morrison, D.A. (in press, as above)) than the strategies implemented in GARLI (14) or RAxML (15). To assess confidence in clades, bootstrapping was performed by executing PRAP-generated command files in PAUP. Using optimized parameters from the likelihood ratchet search, SPR branch swapping was performed on the maximum-likelihood topology for each bootstrapped data matrix, and the proportion of iterations in which a given clade was recovered was mapped onto the maximum-likelihood tree using a strongly modified version of TreeGraph (16) (Müller et al., manuscript in preparation). The latter program was also used to generate SVG trees that can be viewed via the web interface.
Understanding how gene expression patterns vary among gene family members will inform our understanding of evolutionary processes shaping plant gene function and genome structure. Characterization of changing gene expression following gene duplication and speciation events e.g. Ref. (17), will improve as additional plant genomes are sequenced and genome-wide gene expression studies are preformed on a wide range of plant species (18). We aim to advance this research by placing gene expression data within a gene family context. To incorporate expression data into PlantTribes, we downloaded all AFFY expression data and associated descriptions of experiments, tissue, etc. from NASCArrays (19). This has allowed us to link tribes with Arabidopsis genes to a curated expression dataset including 327 experiments conducted on more than 200 tissues and organs, developmental stages and growth conditions. Gene expression data for additional species will be added to future versions of PlantTribes, as an ontology is developed to relate organs and developmental stages across plant species (20–22).
PLANTTRIBES: DATA ACCESS AND RETRIEVAL
All output discussed in the previous section was loaded into a MySQL database with user-searchable CGI scripts. There are four main ways to search within the PlantTribes Database (Figure 2): (i) using a gene ID or annotation term for any of the Arabidopsis, rice, Populus, Medicago or papaya gene models; (ii) using a CDD domain accession ID, name or description; (iii) running a BLAST search on a single sequence or file of user-supplied sequences or (iv) querying the database of tribe characteristics. For example, all tribes with a minimum and/or maximum tribe size for each species or a threshold cumulative gene number can be retrieved with a simple query. HTML formatted search results include hyperlinks to sequence information, domain content and the tribes represented by each hit. All search results have links to the main page for each tribe within the database. Each tribe page includes the following information: unified annotation, stringency, SuperTribe identifier, the number of sequences from each species, all CDD entries for each of the genes in the tribe, a list of the genes in the tribe as well as each gene's tribe identifier at low, medium and high stringency. Tribe stability can be readily examined through comparison of tribe membership at the different stringencies. Tools are also provided to view sequences from other species that have been sorted into each tribe and to view and/or download the sequences, alignments (constructed at both protein and DNA level) and phylogenetic trees in all major formats for each tribe.
Figure 2.
Schematic diagram describing navigation through the PlantTribes database. (A) A user can search by gene, domain, gene ontology, TAIR gene family annotations and tribe size. (B) All search results are linked into (C) a tribe page with information about the tribe including the distribution of tribe sizes at low, medium and high stringency MCL clustering, links to (D) super tribe pages, domain information for all member genes of the tribe, a listing of all genes within the tribe and (E) a download/view area of additional data for each tribe including sequences, alignments, phylogenetic trees and microarray expression data.
THE UTILITY OF PLANTTRIBES FOR GENE FAMILY ANALYSES
An important utility of PlantTribes is the ability to quickly find organism-specific tribes. In poplar (23), the first sequenced tree, we were able to use this feature to identify tribes containing genes that are unique to that species (among those with sequenced genomes; criteria were no hits with e-values better than the 1.0E–10 threshold). These genome-specific gene models were quite distinct with no hits to any other sequences outside of their tribes. Genes with similar sequences, however, were found in the TIGR Plant Transcript Assembly database (e-values < 1.0E–10 in tblastx searches), suggesting that these may be expressed genes rather than annotation artifacts. A similar experimental strategy can be adopted to identify all organism-specific gene families as well as gene families that have been lost in one or more lineages. Beyond the insights gained from the comparisons of the gene families, the PlantTribes database provides a useful scaffold for sorting new sequences. For example, the Floral Genome Project (24–26) has been sorting ESTs into the putative gene families defined by PlantTribes. Each unigene is searched against the fully sequenced plant proteomes using blastx, best hits are recorded and then used to tentatively place each unigene into a tribe. Further evaluation of tribe membership is facilitated by reports for each unigene showing the best hit(s) and the proportion of genes within each tribe with significant blast scores. This process has allowed us to immediately produce classifications of the genes we are finding in our EST data (25–29). We have used the PlantTribes database to identify single copy tribes (genes with just one member from each species) whose memberships were stable across all three stringencies (30). These shared single-copy genes are more abundant than expected by chance, given the frequency of single-copy genes that have resulted from gene death following gene and genome duplication in these lineages in Arabidopsis, rice and Poplar, (30). A similar strategy can be used to produce a list of all of the tribes that are present in only one of the species, i.e. tribes with zero genes in all species but one. Identifying orthologs in EST sets from several basal angiosperms has also allowed us to infer lineage-specific substitution rate variation (31). The database has also aided the identification of paralogous pairs to explore gene duplication through angiosperm history (17) and assess the frequency of expression shifts following gene duplication. In contrast, conserved gene expression in some single-copy genes suggests conserved function throughout angiosperm history (30).
PlantTribes circumscribes objectively defined gene families, but we need to assess the degree to which the MCL clustering algorithm recovers evolutionary complete gene family clades. Using the expansin and MADS-box gene families as exemplars, we mapped tribe assignments at low, medium and high stringencies onto previously reported phylogenies for these two well-studied gene families. We wanted to test the extent to which tribes represent ‘putative’ gene families and investigate whether large tribes recovered at low stringency typically break up at higher stringencies into smaller tribes corresponding to subfamilies. We tested whether there is a strict nested relationship among tribes identified at low, medium and high stringencies, and if so, whether the nested pattern of relationships corresponds to the historical relationships and past gene-duplication events as estimated in phylogenetic analyses. Figure 3A contains the mapping of tribe membership from the three-way clustering to a phylogeny of the expansin superfamily (32). All of the expansin genes reported in the phylogeny from three expansin subfamilies, alpha, beta and expansin-like, are found in only one tribe at low stringency. At medium stringency, the genes are broken into two tribes with one tribe containing the alpha and beta-expansins and the second tribe containing the expansin-like genes. At the highest stringency, the expansin-like alpha and expansin-like beta subfamilies are split from the main tribe containing the alpha and beta expansins.
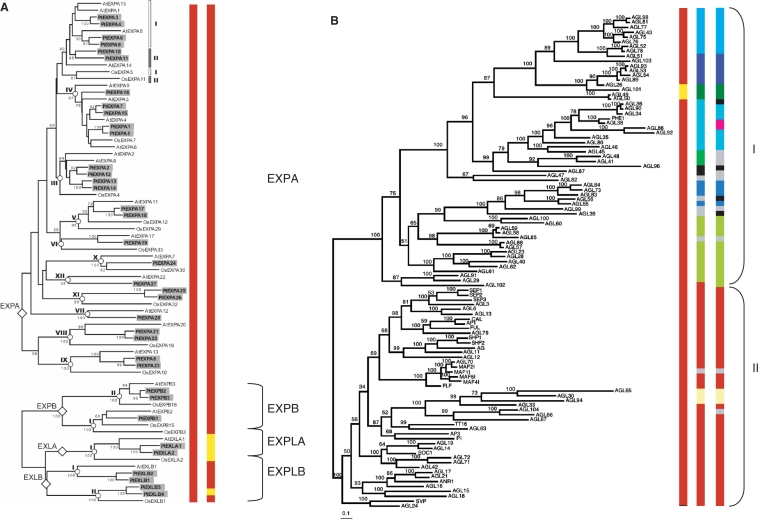
Figure 3.
Tribes with expansin (A) and MADS box genes (B) formed at low, medium and high stringencies in the three-species clustering are mapped onto recently published gene phylogenies (32,33). (A) In the Expansin phylogeny, all genes are found in a single tribe at low stringency. At medium stringency, the genes are broken up into two tribes separating expansin-like A subfamily genes from all others expansin sub-families (tribe containing additional expansin-like genes not included in the original phylogeny). At high stringency, expansins are resolved as two tribes corresponding to the sub-families alpha + beta and expansin-like. (B) The MADS box genes (including type I and II) included in the phylogeny are in two tribes with all genes in one tribe except AGL49 and AGL50. At medium and high stringencies, well-defined clades appear. The type I genes break up into many more tribes than type II genes, which is expected since type I genes are more divergent among themselves. Within the type II genes, AGL65, AGL30, AGL94 are broken out from the main tribe, which is to be expected since this group of genes is highly divergent type II genes.
It would be desirable for tribes to generally correspond to monophyletic clades as was the case in the expansin superfamily. This would allow investigators to download and align sequences from a tribe with confidence that all genes in the alignment share a common ancestor and all extant descendents of that ancestral gene are included in the alignment. However, this is not always the case. Figure 3B contains the mapping of tribe membership from the three-way clustering to a well-accepted Arabidopsis MADS-box gene family phylogeny (33). At higher stringencies, small groups of related genes are peeled off of the low-stringency clusters. As a consequence, the largest tribe identified at high stringency is paraphyletic with some divergent internal clades segregating into distinct tribes. Whereas nearly all type-II MADS-box genes were placed in a single tribe at low stringency, the complete MADS-box gene family (including type-I) was distributed among 14 tribes in the 5-species analysis. Even though the tribes at three stringencies may not always coincide with clades at each hierarchical level, the ability of tribe and supertribe analyses to capture a large number of related genes nevertheless provides an efficient starting point for investigations of gene and gene-family diversification across complete genomes.
CONCLUSIONS AND FUTURE PERSPECTIVES
The PlantTribes database offers a unique and powerful view of plant genomes and evolution. Collaborators working on annotation and interpretation of gene models for the Poplar and papaya genomes found the tribe results to be an invaluable tool for gene family identification and annotation, and our results were highlighted in the recent Poplar genome sequence paper (23). More than 15 other published articles to date have relied on data extracted from PlantTribes including expression divergence following gene duplication, identification of novel functional motifs, identification of gene families for intensive phylogenetic analysis and genome duplication history of basal angiosperms. With many plant genome sequence projects in progress, formal comparative approaches such as PlantTribes will allow researchers to rapidly identify the best gene models, quickly determine errors in the initial annotations, identify new gene families and increase the confidence in the limits and structure of existing gene families.
PlantTribes has been designed for ease of expansion and feature addition. As new genomes are sequenced, or large EST sets generated, PlantTribes will be continuously expanded to include these data. New features being developed presently include (i) a tool for the rapid incorporation of new query sequences into tribe alignments and phylogenies, (ii) connecting the rapidly expanding microRNA database into PlantTribes so that genes that are putative targets of known or predicted miRNAs may be easily found, (iii) expansion of the microarray database to include large-scale array experiments from basal angiosperms and other plants (26) that will facilitate cross-species expression analyses and (iv) synteny-based tools to map genome duplications onto gene-family phylogenies. As the number of sequenced genomes increases rapidly, the continued expansion of the PlantTribes database will facilitate a multitude of genome and gene-family studies, particularly homology-based annotation, genome-scale analysis of multiple gene families, characterization of large gene families and subsets of genes with common domain architectures.
ACKNOWLEDGEMENTS
The authors thank our faculty, postdoctoral and student colleagues in the Floral Genome Project, the Ancestral Angiosperm Genome Project and the poplar and papaya genome projects for their enthusiastic support and use of PlantTribes through its initial stages of development. We would like to thank Hong Ma, John Carlson and Victor Albert for invaluable discussions of the biological implications of PlantTribes. Finally, we thank Josh Marion, Tony Orenga, Severn Everett, Kevin Beckmann, Anthony Carroll and Erik Wolcott for their assistance in the development of portions of the PlantTribes database and web interface. This work was funded by National Science Foundation (DEB 0115684 to C.W.D. and J.L.-M., DEB 0638595 to C.W.D. and J.L.-M, and DBI-0501890 to C.W.D.). K.F.M. was supported by a scholarship from the Deutsche Telekom Stiftung. Funding to pay the Open Access publication charges for this article was provided by NSF DEB 0638595.
Conflict of interest statement. None declared.
REFERENCES
- 1.Van Dongen S. Technical Report INS-R0010. 2000. A cluster algorithm for graphs. [Google Scholar]
- 2.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong Q, Schlueter SD, Brendel V. PlantGDB, plant genome database and analysis tools. Nucleic Acids Res. 2004;32:D354–D359. doi: 10.1093/nar/gkh046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudd S. openSputnik – a database to ESTablish comparative plant genomics using unsaturated sequence collections. Nucleic Acids Res. 2005;33:D622–D627. doi: 10.1093/nar/gki040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartmann S, Lu D, Phillips J, Vision TJ. Phytome: a platform for plant comparative genomics. Nucleic Acids Res. 2006;34:D724–D730. doi: 10.1093/nar/gkj045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Childs KL, Hamilton JP, Zhu W, Ly E, Cheung F, Wu H, Rabinowicz PD, Town CD, Buell CR, et al. The TIGR Plant Transcript Assemblies database. Nucleic Acids Res. 2007;35:D846–D851. doi: 10.1093/nar/gkl785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 8.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller KF. PRAP – computation of Bremer support for large data sets. Mol. Phylogenet. Evol. 2004;31:780–782. doi: 10.1016/j.ympev.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Swofford DL. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4. [Google Scholar]
- 13.Nixon KC. The Parsimony Ratchet, a new method for rapid parsimony analysis. Cladistics. 1999;15:407–414. doi: 10.1111/j.1096-0031.1999.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 14.Zwickl DI. Ph.D. Thesis. The University of Texas at Austin; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. [Google Scholar]
- 15.Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 16.Müller J, Müller K. TreeGraph: automated drawing of complex tree figures using an extensible tree description format. Mol. Ecol. Notes. 2004;4:786–788. [Google Scholar]
- 17.Duarte JM, Cui LY, Wall PK, Zhang Q, Zhang XH, Leebens-Mack J, Ma H, Altman N, dePamphilis CW. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol. Biol. Evol. 2006;23:469–478. doi: 10.1093/molbev/msj051. [DOI] [PubMed] [Google Scholar]
- 18.Shen L, Gong J, Caldo RA, Nettleton D, Cook D, Wise RP, Dickerson JA. BarleyBase – an expression profiling database for plant genomics. Nucleic Acids Res. 2005;33:D614–D618. doi: 10.1093/nar/gki123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S. NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 2004;32:D575–D577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buzgo M, Soltis DE, Soltis PS, Ma H. Towards a comprehensive integration of morphological and genetic studies of floral development. Trends Plant Sci. 2004;9:164–173. doi: 10.1016/j.tplants.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Ilic K, Kellogg EA, Jaiswal P, Zapata F, Stevens PF, Vincent LP, Avraham S, Reiser L, Pujar A, et al. The plant structure ontology, a unified vocabulary of anatomy and morphology of a flowering plant. Plant Physiol. 2007;143:587–599. doi: 10.1104/pp.106.092825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pujar A, Jaiswal P, Kellogg EA, Ilic K, Vincent L, Avraham S, Stevens P, Zapata F, Reiser L, et al. Whole-plant growth stage ontology for angiosperms and its application in plant biology. Plant Physiol. 2006;142:414–428. doi: 10.1104/pp.106.085720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 24.Soltis DE, Soltis PS, Albert VA, Oppenheimer DG, dePamphilis CW, Ma H, Frohlich MW, Theissen G. Missing links: the genetic architecture of flowers [correction of flower] and floral diversification. Trends Plant Sci. 2002;7:22–31. doi: 10.1016/s1360-1385(01)02098-2. dicussion 31–24. [DOI] [PubMed] [Google Scholar]
- 25.Albert VA, Soltis DE, Carlson JE, Farmerie WG, Wall PK, Ilut DC, Solow TM, Mueller LA, Landherr LL, et al. Floral gene resources from basal angiosperms for comparative genomics research. BMC Plant Biol. 2005;5:5. doi: 10.1186/1471-2229-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soltis DE, Ma H, Frohlich MW, Soltis PS, Albert VA, Oppenheimer DG, Altman NS, Depamphilis C, Leebens-Mack J. The floral genome: an evolutionary history of gene duplication and shifting patterns of gene expression. Trends Plant Sci. 2007;12:358–367. doi: 10.1016/j.tplants.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 27.Carlson JE, Leebens-Mack JH, Wall PK, Zahn LM, Mueller LA, Landherr LL, Hu Y, Ilut DC, Arrington JM, et al. EST database for early flower development in California poppy (Eschscholzia californica Cham., Papaveraceae) tags over 6000 genes from a basal eudicot. Plant Mol. Biol. 2006;62:351–369. doi: 10.1007/s11103-006-9025-y. [DOI] [PubMed] [Google Scholar]
- 28.Kim S, Soltis PS, Wall K, Soltis DE. Phylogeny and domain evolution in the APETALA2-like gene family. Mol. Biol. Evol. 2006;23:107–120. doi: 10.1093/molbev/msj014. [DOI] [PubMed] [Google Scholar]
- 29.Duarte JM, Wall PK, Zahn LM, Leebens-Mack JH, dePamphilis CW. Taxon. Utility of Amborella trichopoda and Nuphar advena ESTs for phylogeny and comparative sequence analysis. (in press) [Google Scholar]
- 30.Leebens-Mack JH, Wall K, Zheng Z, Oppenheimer D, dePamphilis CW. Developmental Genetics of the Flower. London: Elsevier Limited; 2006. [Google Scholar]
- 31.Cui L, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, Soltis PS, Carlson JE, Arumuganathan K, et al. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006;16:738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampedro J, Carey RE, Cosgrove DJ. Genome histories clarify evolution of the expansin superfamily: new insights from the poplar genome and pine ESTs. J. Plant Res. 2006;119:11–21. doi: 10.1007/s10265-005-0253-z. [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Castilla LP, Alvarez-Buylla ER. Adaptive evolution in the Arabidopsis MADS-box gene family inferred from its complete resolved phylogeny. Proc. Natl Acad. Sci. USA. 2003;100:13407–13412. doi: 10.1073/pnas.1835864100. [DOI] [PMC free article] [PubMed] [Google Scholar]