Abstract
WormBase (www.wormbase.org) is the major publicly available database of information about Caenorhabditis elegans, an important system for basic biological and biomedical research. Derived from the initial ACeDB database of C. elegans genetic and sequence information, WormBase now includes the genomic, anatomical and functional information about C. elegans, other Caenorhabditis species and other nematodes. As such, it is a crucial resource not only for C. elegans biologists but the larger biomedical and bioinformatics communities. Coverage of core areas of C. elegans biology will allow the biomedical community to make full use of the results of intensive molecular genetic analysis and functional genomic studies of this organism. Improved search and display tools, wider cross-species comparisons and extended ontologies are some of the features that will help scientists extend their research and take advantage of other nematode species genome sequences.
PROGRESS
Ongoing Caenorhabditis elegans research has led to increases in existing data sets such as RNAi and expression patterns and the development of novel data types including mass spectrometry. As comparative genomics has become an important research approach, we have incorporated relevant genomes as they have become available. In addition to the existing Caenorhabditis remanei and updated Caenorhabditis briggsae assembly browsers a version of the Brugia malayi genome (1) is also available through the website. The volume and variety of functional data available through WormBase continues to grow. WormBase tries to make this readily available and searchable in several ways. A manually curated summary is available for 4780 of the most heavily researched genes, an increase of 423 since last year. Standardized ontologies provide consistent annotation for all genes, especially useful for those lacking detailed experimental evidence. Furthermore, these ontologies by extension provide a mechanism for users to bootstrap into understanding gene function in related but less well-studied species. The most widely used of these is the Gene Ontology (GO) (2). GO terms are added to genes based on manual curation of the literature and by inference from protein domains and phenotypes caused by mutation or RNAi. The Gene Ontology, along with ontologies of phenotype descriptions and anatomy terms, is browsable through a newly developed ontology viewer. Parent and child terms are expandable and links to relevant genes or other data types shown.
WEBSITE OUTLINE
The front page of WormBase provides users a convenient mechanism to search all contents of the database. Most searches centre on genes; gene pages act as a summary or portal page, displaying information such as gene identifiers, exon structure, experimental and functional data, similarity to other genes and gene families and reagents available for researchers. Most data types link to specialized report pages containing further detailed information. All genes for which the sequence is known are linked to a GBrowse-driven genome browser, which has a large number of configurable tracks allowing users to customize the data they see. Powerful search tools such as a BioMart (3) implementation and BLAST server allow users to easily mine WormBase and query relevant features by sequence identity.
GENE STRUCTURE AND GENOME SEQUENCE CURATION
Caenorhabditis elegans gene structures have been manually curated for over a decade; still, we are far from having a guaranteed ‘gold standard’ gene set. More than 1500 changes were still necessary in the past year based on new evidence and analysis methods. These improvements mean that over a third of C. elegans protein-coding genes are now fully covered from ATG to stop by transcript data. The importance of non-coding RNA genes is becoming more apparent (4) and WormBase has made significant efforts to incorporate over 5500 published novel RNA genes (5–8). The genome sequence is also still subject to change based on user-provoked re-inspection of the original sequencing data.
MASS SPECTROMETRY
The first C. elegans mass spectrometry data consisting of 79 844 mass-spec peptides has been added to WormBase (9) (G Merrihew and Lukas Reiter, personal communication). All of these peptides are mapped to proteins and the genome and are an extremely useful aid in the confirmation of the gene structures. There are 120 genes with no transcript support that have mass spectrometry support.
COMPARATIVE GENOMICS
There have been several significant improvements to the comparative genomics resources in WormBase. Building on the use of Ensembl technology (10,11) WormBase now displays orthologous genes found in many of the vertebrate species included in Ensembl such as human (11 801) and mouse (12 141). There are several other methods for determining orthologous relationships between genes in different species and the different methods do not always agree. Rather than selecting a single method of ortholog determination, WormBase has chosen to represent a selection of methods and let the user decide. To this end, homology groups are also included from InParanoid (12), NCBI-KOGS (13), TreeFam (14), OrthoMCL (15) as well as one-off analyses (16) and individual user contributions.
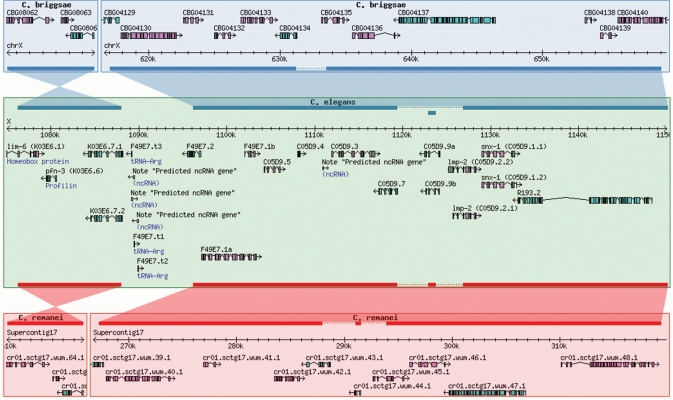
The Compara system that is used to calculate these orthologs and paralogs also determines syntenic regions between the more closely related Caenorhabditis species (currently elegans, briggsae, remanei and brenneri). These alignments are used to drive a recently developed synteny viewer (Figure 1), which is accessible via a new link in the main navigation bar at the top of each page. Users can scroll and zoom around the genome as with the main Genome Browser, change which genome is to act as the reference and follow links to genes shown.
Figure 1.
Syntenic alignments of C. elegans, C. briggsae and C. remanei.
We now manually curate C. briggsae gene predictions in response to user queries. Genome-wide experiments in C. briggsae are also being included with the addition of 42 500 SNPs identified by the Washington University SNP Research Facility which are used to generate a genetic map.
To make it easier for users to explore differences in gene structure between species, we have added a new track to the Genome Browser showing the alignment of cDNA data from other Caenorhabditis species to the elegans genome. This complements the existing tracks where cDNAs from C. elegans itself are separated from those of all other nematodes. This track gives a quick overview of the level of conservation between the various Caenorhabditis species. The species from which the cDNA derives is indicated as a mouse-over tool tip. As the genome sequences of the other Caenorhabditis are annotated and integrated, genome browsers will be established with similar tracks of cDNA alignments where possible. The latest species added to the genome browser is Brugia malayi (1), sequenced by TIGR and we expect to be adding an updated C. remanei genome plus new browsers for C. brenneri and Caenorhabditis japonica (17) in the near future. In addition, WormBase is actively working with the Nematode Genome Annotation Assessment Project (nGASP) (www.wormbase.org/wiki/index.php/Gene_Prediction) to determine, display and maintain a canonical gene set for each species.
GENE EXPRESSION
There are several methods used to investigate a gene's expression that are displayed through WormBase (Figure 2). The extent of microarray data continues to grow with another 14 papers (211 experiments) added (18–20). SAGE data provides large-scale gene expression analysis and can indicate relative levels of mRNAs. The SAGE data in WormBase now includes all data sets currently available for C. elegans and the display of these sequences has been improved so that the orientation and frequency count for each tag is easily viewable.
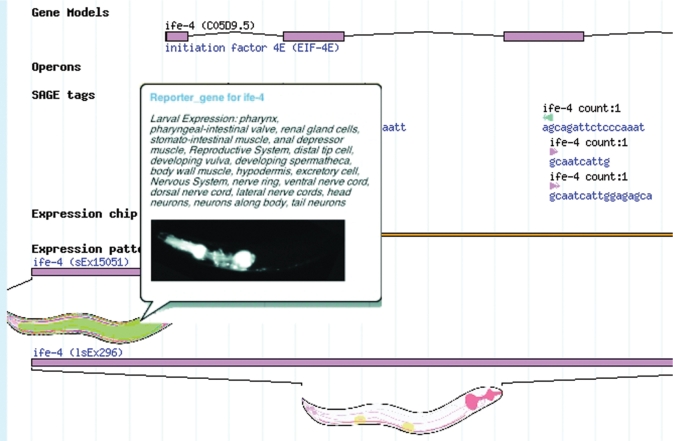
Figure 2.
Expression details of ife-4 shown in genome browser. Cartoon worms show approximate expression location. Mouse over pop-up window (left-center box) gives more details and original image. SAGE tags shown as blue/pink triangles with tag sequence and count. Sequences used in microarray experiments shown as horizontal, orange bar which can be clicked for more details. Gene exons displayed as pink boxes.
In parallel to the improved expression pattern display the anatomy ontology has been extended in breadth and depth. With this controlled vocabulary, it is now possible to link gene expression and phenotypes to distinct anatomical features in the worm.
GENE INTERACTIONS AND REGULATION
Determining how genes interact and regulate each other is vital for understanding how they function in the context of the system, cell or animal. To capture this type of data, WormBase has improved the granularity of the interaction data captured so that interactions between specific allelic variants of genes can be described and phenotypes shown. There are a wide variety of experimental methods used to investigate interactions such as Yeast one- (21), two- (22) and three-hybrid (23) and RNAi in mutant backgrounds. These involve several molecule types, including DNA, RNA and protein. The 5431 experimentally determined, C. elegans interactions are both manually (377) and automatically [5054—via Textpresso (24)] curated with an emphasis on including large-scale data sets where possible. Over 17 000 predicted interactions based on orthologous interactions are also displayed (25). The wide range of interactions displayed through the website is greatly enhanced by the incorporation of the molecular interaction network browser—N-Browse, developed by Kris Gunsalus’ group at NYU (Figure 3) (www.gnetbrowse.org).
Figure 3.
N-Browse can be accessed via the icon on the Gene page (inset). Gene information (left panel) is taken from WormBase. Each gene in the main network window can be clicked to expand the network. Interaction types are colour-coded and can be switched dynamically on or off with the check boxes (lower panel). Search and navigation tools are provided in the top bar.
GENETIC DATA
The genetic map of C. elegans and well-described alleles are important resources for researchers. The latest WormBase survey shows that community users are expressing an overwhelming interest in detailed allele-specific information, both molecular and phenotypic (see Phenotype section below). All sequenced alleles described in papers published since 2001 are now available in WormBase. Where details of the molecular nature of a change are available they are recorded and located on the genome sequence. As gene predictions are still actively curated it is possible that a gene affected by an allele may change so the connections between genes and alleles are updated for every release. The impact on the protein of the allele changes are also dynamically inferred from the DNA change. Alleles published prior to 2001 are now being curated. We continue to incorporate the disruption alleles systematically produced by the National Bioresource Project for the Experimental Animal ‘Nematode’, the C. elegans Gene Knockout Consortium and NemaGENETAG (elegans.imbb.forth.gr/nemagenetag/) projects. The Variation Summary, which displays Alleles and SNPs, has been expanded to display curated molecular information details. This includes sequence context of the variation, conceptual translations of missense and nonsense alleles and images that show the position of the variation in relation to genomic features like gene models, and translated features such as motifs and BLASTP homologies.
The Genetic Map display has been updated and now uses GBrowse (the same interface as the Genome Browser) technology. The familiar interface is user-friendly and can easily be configured. It displays all objects that have a genetic map position including landmark genes, chromosomal rearrangements, interpolated genes, alleles and SNPs. The method of calculating interpolated map positions has been improved. Previously, map positions were calculated assuming an equal recombination rate across the chromosome, whereas the new version interpolates between two neighbouring markers allowing for differences in recombination rates along a chromosome.
PHENOTYPE
Over the past 2 years, we have spent considerable effort developing a Phenotype Ontology in order to have a controlled language for phenotype terms. Our most recent effort has focused on implementing this ontology to link accurate and detailed phenotype information to genetic variation objects (e.g. alleles, RNAi and transgenes) associated with genes. As of the September 2007 database release (WS184), the number of phenotypes connected to variation objects has increased 290% (from 71 972 to 281 360 total phenotype connections: 6555 phenotype connections for 3510 alleles, 274 766 for 65 026 RNAi objects and 39 for 7 transgenes). The curation of phenotype objects necessarily occurs in parallel with the ongoing development of the Phenotype Ontology to which we have added 270 new terms since last year. This ongoing development is crucial for phenotype information to reflect the complexity and richness of experimental results. Users have emphasized that detailed allele descriptions are highly valued so phenotype annotation and concurrent Phenotype Ontology development will remain a high priority in the upcoming years.
IMPROVED SEARCHING
We continue to refine the user interface of WormBase to make searches fast and intuitive. The main search box on the home page now offers an optional ‘autocomplete’ facility, which uses a separately indexed database to offer suggestions as users type their query. The list can be navigated by mouse or cursor keys to give rapid access to the object being searched for. The presence of an autocomplete ‘hit’ lets users quickly see search terms that will yield positive results.
WormMart, the WormBase-specific implementation of the BioMart system (3), has had its user interface revamped. Instead of the page-by-page approach of the original version, a selection of expandable and collapsible panels is used to select, filter and export data. To help users explore this complex facility, several examples have been included in the WormBase, Wiki based, FAQ (www.wormbase.org/wiki/index.php/FAQs#WormMart_questions).
Our BLAST service has been extended to include C. elegans, C. briggsae, C. remanei and C. brenneri sequences. A new karyotype image displays the genomic location of positive results, and inline images display the genomic context of the high-scoring sequence pairs.
COMMUNITY INTERACTION
In response to our 2005 User Survey, we have added two features to encourage community interactions. We established the WormBase Wiki, a publicly accessible and editable segment of WormBase. Every gene has a Wiki page, which can be edited by users and we intend to make these comments visible on the gene page. We are also using the Wiki to document WormBase Standard Operating Procedures and Frequently Asked Questions (FAQ). In collaboration with WormAtlas (www.wormatlas.org) and WormBook (26), we launched the Worm Community Forum, a public bulletin board system with active discussion on a variety of topics related to C. elegans biology and research. Another survey was carried out this year which will guide future development of the database and website.
DATABASE ACCESS AND DISTRIBUTION
The primary means of accessing the WormBase data remains through the main website (www.wormbase.org). A WormBase maintained UK mirror (http://wormbase.sanger.ac.uk) has been established to complement the existing European mirrors run by non-WormBase groups in Marseille (http://crfb-3.univ-mrs.fr/) and Crete (http://imbb.wormbase.org). It has been made much easier for individuals to set up their own version of the site through the introduction of WormBase Virtual Machines, software packages that contain all databases and software driven by a guest operating system. When opened in a free application called VMPlayer, the provided guest operating system opens in a window, starts all necessary services and databases, followed by WormBase in a new browser window. This technology is now being used to drive the archival ‘frozen’ releases of WormBase that are maintained as reference points intended for use in genome wide analyses (e.g. http://ws170.wormbase.org).
An assortment of flat files is available for download including genome features in GFF2 and GFF3 format. Commonly requested data sets such as lists of genetic interactions, best blastp hits and intergenic sequences are amongst others. The ACeDB database files are also still distributed.
Much of the sequence based data collated and annotated by WormBase is also available through the Ensembl website (www.ensembl.org/Caenorhabditis_elegans/index.html.) This is updated for every frozen release.
ACKNOWLEDGEMENTS
WormBase is supported by grant P41-HG02223 from the US National Human Genome Research Institute and the British Medical Research Council. P.W.S. is an investigator with the Howard Hughes Medical Institute. Funding to pay the Open Access publication charges for this article was provided by grant P41-HG02223 from the US National Human Genome Research Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.TIGR. Brugia Malayi Genome Sequencing Project. 2005. [cited; Available from: http://www.tigr.org/tdb/e2k1/bma1/.
- 2.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.BioMart. [cited; Available from: http://www.biomart.org/].
- 4.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 5.Zemann A, et al. Evolution of small nucleolar RNAs in nematodes. Nucleic Acids Res. 2006;34:2676–2685. doi: 10.1093/nar/gkl359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wachi M, et al. Isolation of eight novel Caenorhabditis elegans small RNAs. Gene. 2004;335:47–56. doi: 10.1016/j.gene.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Deng W, et al. Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis, and expression. Genome Res. 2006;16:20–29. doi: 10.1101/gr.4139206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 9.Husson SJ, et al. Impaired processing of FLP and NLP peptides in carboxypeptidase E (EGL-21)-deficient Caenorhabditis elegans as analyzed by mass spectrometry. J. Neurochem. 2007;102:246–260. doi: 10.1111/j.1471-4159.2007.04474.x. [DOI] [PubMed] [Google Scholar]
- 10.Hubbard TJ, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potter SC, et al. The Ensembl analysis pipeline. Genome Res. 2004;14:934–941. doi: 10.1101/gr.1859804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Stoeckert C.J., Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillier LW, et al. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 2007;5:e167. doi: 10.1371/journal.pbio.0050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sternberg P. Genome sequence of additional Caenorhabditis species: enhancing the utility of C. elegans as a model organism. 2003. http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/C_remaneiSEQ.pdf.
- 18.Kirienko NV, Fay DS. Transcriptome profiling of the C. elegans Rb ortholog reveals diverse developmental roles. Dev. Biol. 2007;305:674–684. doi: 10.1016/j.ydbio.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer JN, et al. Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome Biol. 2007;8:R70. doi: 10.1186/gb-2007-8-5-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welker NC, Habig JW, Bass BL. Genes misregulated in C. elegans deficient in Dicer, RDE-4, or RDE-1 are enriched for innate immunity genes. RNA. 2007;13:1090–1102. doi: 10.1261/rna.542107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deplancke B, et al. A gateway-compatible yeast one-hybrid system. Genome Res. 2004;14:2093–2101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walhout AJ, Boulton SJ, Vidal M. Yeast two-hybrid systems and protein interaction mapping projects for yeast and worm. Yeast. 2000;17:88–94. doi: 10.1002/1097-0061(20000630)17:2<88::AID-YEA20>3.0.CO;2-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B, et al. Yeast three-hybrid system to detect and analyze RNA-protein interactions. Methods Enzymol. 2000;318:399–419. doi: 10.1016/s0076-6879(00)18066-8. [DOI] [PubMed] [Google Scholar]
- 24.Muller HM, Kenny EE, Sternberg PW. Sternberg, Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong W, Sternberg PW. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311:1481–1484. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
- 26.Girard LR, et al. WormBook: the online review of Caenorhabditis elegans biology. Nucleic Acids Res. 2007;35:D472–D475. doi: 10.1093/nar/gkl894. [DOI] [PMC free article] [PubMed] [Google Scholar]