Abstract
G-quadruplex motifs in the RNA play significant roles in key cellular processes and human disease. While sequences capable of forming G-quadruplexes in the pre-mRNA are involved in regulation of polyadenylation and splicing events in mammalian transcripts, the G-quadruplex motifs in the UTRs may help regulate mRNA expression. GRSDB2 is a second-generation database containing information on the composition and distribution of putative Quadruplex-forming G-Rich Sequences (QGRS) mapped in ∼29 000 eukaryotic pre-mRNA sequences, many of which are alternatively processed. The data stored in the GRSDB2 is based on computational analysis of NCBI Entrez Gene entries with the help of an improved version of the QGRS Mapper program. The database allows complex queries with a wide variety of parameters, including Gene Ontology terms. The data is displayed in a variety of formats with several additional computational capabilities. We have also developed a new database, GRS_UTRdb, containing information on the composition and distribution patterns of putative QGRS in the 5′- and 3′-UTRs of eukaryotic mRNA sequences. The goal of these experiments has been to build freely accessible resources for exploring the role of G-quadruplex structure in regulation of gene expression at post-transcriptional level. The databases can be accessed at the G-Quadruplex Resource Site at: http://bioinformatics.ramapo.edu/GQRS/.
INTRODUCTION
The G-rich polynucleotide molecule can repeatedly fold on itself to form a unimolecular quadruplex structure consisting of stacked G-tetrads, which are square co-planar arrays of four guanine bases each (1). Although G-quadruplexes can also be formed by association of two or four molecules, the present work focuses only on the unimolecular quadruplexes which are more likely to be encountered in physiological conditions (2,3).
G-quadruplexes have come into the limelight in recent years, especially because of increasing indication for their diverse roles in key cellular processes, human disease and as targets for therapy (4–10). Production of G-quadruplexes has been shown to occur cotranscriptionally in the G-rich complementary DNA strands (11). Formation of RNA G-quadruplex structures in vivo has also been demonstrated (12). In fact, RNA is more likely to form stable G-quadruplexes than DNA in vivo (13,14). G-quadruplex motifs in the RNA have been shown to play significant roles in mRNA turnover (4) and FMRP binding (15). Genes containing FMRP-binding sites may be regulated by a common pathway. At least two such genes have been suggested to be involved in autism (16). We have previously shown that interaction of a G-rich Sequence (GRS) with hnRNPH/H′ can modulate 3′end processing of mammalian pre-mRNAs (17–19). Recently, Furger and coworkers have also found that 3′end processing of melanocortin receptor 1 is regulated by interaction of two G-rich elements with hnRNPH/H′ (20). We have determined, using the QGRS Mapper software program that we had developed earlier (21), that GRS in the above studies are potentially capable of forming stable G-quadruplexes. The hnRNPs H/H′ and F that bind to GRS are known to regulate polyadenylation and splicing events in mammalian transcripts (22–24). The hnRNP A1, which is found in alternative splicing reactions, also has a demonstrated affinity for the G-quadruplex structure (25). G-rich motifs that may fold into quadruplexes in the vicinity of RNA-processing sites act as regulators by interacting with hnRNP A1, H/H′ or F proteins (17–19,24,26,27). The majority of human genes are known to undergo alternative polyadenylation (28), or alternative splicing (29). The role of quadruplex structure in regulating RNA processing, which is an essential component of differential gene expression, needs to be explored.
Prevalence of G-quadruplexes in the human genome has been established (30,31). In a recent study, gene function was found to be associated with potential for G-quadruplex formation (32). However, there is a paucity of systematic studies focusing on the analysis of G-quadruplex motifs near RNA processing sites of the genes, especially that are alternatively processed. Genes that contain G-quadruplex forming sequences are likely to be regulated via special mechanisms (32). Our group has been interested in studying the role of G-quadruplexes in regulation of gene expression at post-transcriptional level. We have adopted a bioinformatics approach to study composition and patterns of G-quadruplexes in pre-mRNA and mRNA sequences.
We had previously built GRSDB, a database of mapped G-quadruplex sequences in selected alternatively processed human and mouse genes (33). GRSDB2 is a second-generation database and contains information on composition and distribution of putative Quadruplex-forming G-Rich Sequences (QGRS) mapped in a large number of eukaryotic pre-mRNA sequences, many of which are alternatively processed (alternatively spliced or alternatively polyadenylated). The data stored in the GRSDB2 is based on computational analysis of NCBI Entrez Gene entries and their corresponding annotated genomic nucleotide sequences of RefSeq/GenBank. GRSDB2 has been built with a new and much improved version of QGRS Mapper program (21). It contains data from ∼29 000 eukaryotic genes from other organisms in addition to human and mouse. The data model of GRSDB2 is different than the first version in that it is centered around Entrez Gene rather than solely GenBank/RefSeq nucleotide entries. The search module has been greatly enhanced, making it possible to generate complex queries to search the database with a wide variety of parameters including Gene Ontology terms. The user may select subsets of genes from a query and perform further computations on these genes through a ‘Workbench’. It is also possible to define the composition and size of G-quadruplexes to be displayed by applying a variety of filters through the ‘options’ menu. The ‘Gene View’, ‘Data View’ and a highly interactive ‘Graphic View’ for individual database entries have been significantly enhanced with several additional computational capabilities and links. The data can now be exported into Excel for further analysis. In addition, we have added a ‘Sequence View’, which displays mapped G-quadruplexes in the context of pre-mRNA sequence.
G-quadruplexes in the mRNA can influence translation initiation (34) as well as repression (35). Recently, a G-quadruplex in the 5′-UTR (untranslated region) of NRAS proto-oncogene mRNA was found to inhibit its translation (14). This study also found G-quadruplexes in the 5′-UTR of many other genes. The UTRs of mRNAs contain motifs that are vital for regulation of post-transcriptional gene expression. Much attention has been paid to study the composition of regulatory RNA motifs and mechanism of their interactions with the cellular machinery (36). Our preliminary bioinformatics studies have found notable frequencies of G-quadruplex motifs in the 5′- as well as 3′- UTRs of mammalian mRNAs. More detailed studies are needed to investigate the role of UTR G-quadruplex structure in regulating post-transcriptional gene expression. We have developed a new database, GRS_UTRdb, which contains information on the composition and distribution patterns of putative Quadruplex forming GRS in the 5′- and 3′-UTRs of eukaryotic mRNA sequences. The data stored in the GRS_UTRdb is based on computational analysis of NCBI Entrez Gene entries and their corresponding annotated nucleotide sequences of RefSeq/GenBank. The computations were performed with the help of an extension of the existing QGRS Mapper program (20).
Both the GRSDB2 and GRS_UTRdb databases can be accessed at the G-Quadruplex Resource Site at: http://bioinformatics.ramapo.edu/GQRS/. The goal of these experiments has been to build resources for exploring the role of G-quadruplex structure in regulation of gene expression at post-transcriptional level. Researchers will find both the websites to be user-friendly along with comprehensive help sections as well as context-specific help where it is needed. Investigators interested in the functional relevance of G-quadruplex structure, in particular its role in regulating the gene expression at post-transcriptional level, will find both the databases to be of great value. While GRSDB2 is useful for studying G-quadruplexes near RNA-processing sites, particularly in alternatively processed pre-mRNAs, GRS_UTRdb offers a resource for investigating G-quadruplexes in the untranslated regions of mRNA. Both the websites allow a comprehensive large-scale analysis as well as detailed studies in individual genes.
G-QUADRUPLEX MOTIF
The G-quadruplex motif may be written GxNaGxNbGxNcGx, namely, four guanine groups of equal size (which we call G-groups) interspersed by three arbitrary nucleotide sequences called loops. The size of each G-group corresponds to the number of stacked G-tetrads forming the quadruplex structure. We have previously described the G-quadruplex motif in more detail (21).
The potential of G-quadruplex to influence gene expression relies on the stability of the structure. Stability of the G-quadruplex is considered to be linked to its loop lengths and the number of G-tetrads in the folded structure (37–39). While quadruplexes with at least three G-tetrads have been accepted as stable structures, two G-tetrad quadruplexes are not uncommon (40,41). In fact, a stable two G-tetrad RNA G-quadruplex that is capable of significantly influencing gene expression in vivo has recently been reported (12). Lower stability, in fact may allow more sensitive control of gene expression (12). Two G-tetrads, although relatively lower in stability, are expected to be far more prevalent in the genomes as compared to the three G-tetrads.
METHODS
GRSDB2 and GRS_UTRdb are relational databases developed with MySQL and store non-redundant data. Interfaces for the databases were built using PHP and Java. The databases have been populated with the help of custom software developed previously by us to analyze NCBI Entrez Gene entries (21). The QGRS are mapped within the relevant gene sequence and assigned a computed value, called a G-score, which rewards those sequences deemed more likely to form a stable complex (21).
Structure and features of GRSDB2
GRSDB2 contains information on the composition and distribution of QGRS mapped in the eukaryotic pre-mRNA sequences.
We have made an effort to include all possible G-quadruplexes. Users may search for QGRS containing G-groups of 2, 3 or more. Also, the length of QGRS and loop size are search parameters that the user may set.
There are two categories of QGRS that are regularly used in GRSDB2: (30,2) refers to QGRS at most 30 nt long and having at least 2 G's per G-group, while (45,3) refers to QGRS at most 45 nt long and having at least 3 G's per G-group.
The overall statistics for the database are shown in Table 1.
Table 1.
Statistics for GRSDB2
| Organism | Number of genes | Alternatively spliced | Averagegene size | Average number of products | Number of (30,2) QGRS | Number of (45,3) QGRS |
|---|---|---|---|---|---|---|
| Homo sapiens | 10 475 | 3197 (30.5%) | 61 401 | 1.54 | 2 391 014 | 196 949 |
| Mus musculus | 2008 | 421 (21%) | 52 715 | 1.33 | 371 809 | 31 108 |
| Drosophila melanogaster | 12 223 | 3018 (24.7%) | 5361 | 1.46 | 190 325 | 7830 |
| Rattus norvegicus | 1477 | 37 (2.5%) | 7907 | 1.04 | 39 650 | 2948 |
| Caenorhabditis elegans | 3054 | 1085 (35.5%) | 4478 | 1.54 | 20 194 | 309 |
| Gallus gallus | 41 | 0 (0%) | 19 878 | 1 | 2349 | 211 |
| Bos taurus | 3 | 0 (0%) | 8410 | 1 | 201 | 34 |
| Danio rerio | 7 | 0 (0%) | 15 912 | 1 | 141 | 3 |
| Total | 29 288 | 7758 | 3 015 683 | 239 392 |
Queries may be performed using a variety of search fields. Several fields for gene identifiers are provided: GeneID, Gene Symbol, Gene Name, Aliases, GI and Accession Number. Since alternatively processed genes are a focus of the database, the user may look for genes with a specified number of products and poly(A) signals. The user may also specify which organism(s) to consider.
A significant feature of GRSDB2 is the ability it affords the user to look for correlations of occurrences of G-quadruplexes with gene ontology terms. Queries can specify gene ontology function, process or component. The GO terms are an example of a search field for which the user is not required to exactly match the database entry. Instead, searches may be done for which one or more fields start with, end with or contain the query value.
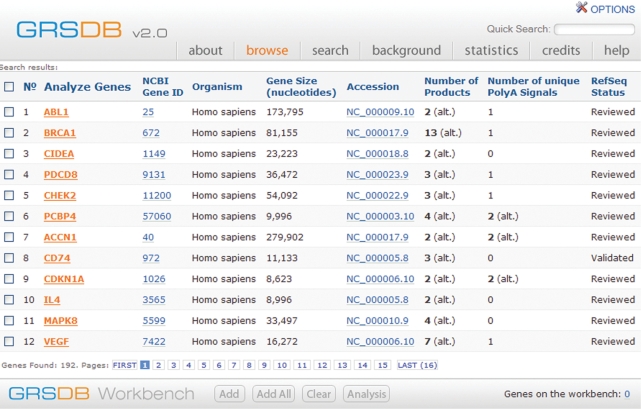
The columns of the query results page consist of the Gene Name, GeneID, Organism, Gene Size, Accession Number, Number of Products and Number of Poly(A) Signals (Figure 1). The results may be sorted on any of these columns, with Gene Name the default sort field. The RefSeq status of each entry is also listed in the table.
Figure 1.
GRSDB2 Query Results Page. Results of a query for alternatively spliced human or rat genes involved in apoptosis. The results may be sorted by clicking the header of any column. At the bottom of the screen, there are four controls allowing the user to add and clear genes from a ‘Workbench’. Several programs are provided on the ‘Workbench’ for further analysis of QGRS from any set of genes in the database.
The user may analyze sets of genes by putting them into a ‘Workbench’, where a variety of programs are available to study QGRS distribution patterns for that particular gene set. There are four controls at the bottom of the results page for working with the ‘Workbench’. The user may add marked genes, add all genes from the query, clear all genes from the ‘Workbench’ or analyze the genes on the ‘Workbench’.
There are five programs available on the ‘Workbench’. One program reports various statistics of the selected genes similar to the statistics page for the entire database. There are two programs [one for (30,2) QGRS, the other for (45,3) QGRS] summarizing the distribution of QGRS with respect to location in exons, introns and near poly(A) signals (which is defined to be within 200 nt). Additionally, there are two programs showing the distribution of G-scores for the selected genes. The user is given the opportunity to export the output of any of these programs to Excel for further analysis.
On the results page, the user may select a particular gene for analysis by clicking on an entry in the Gene Name column. GRSDB2 has five interfaces for viewing information about QGRS: Gene View, Data View (no overlaps), Data View (with overlaps), Sequence View, Graphic View.
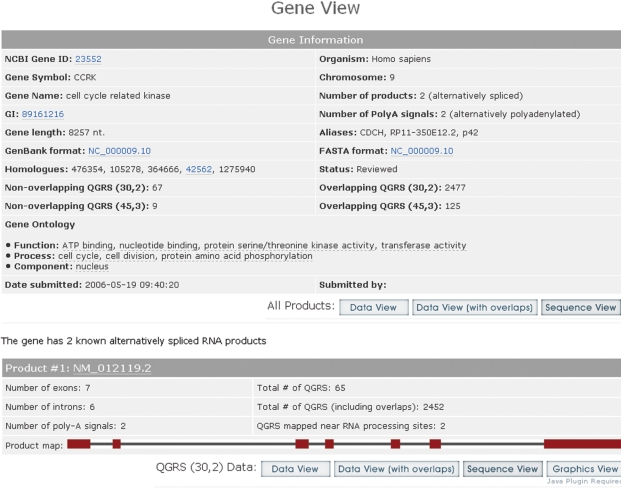
The Gene View has a table presenting basic information on the properties of the gene, the number of QGRS found, and the gene ontology terms associated with the gene (Figure 2). For each alternatively spliced product a map of the exon and intron structure is given together with QGRS information for that product.
Figure 2.
GRSDB2 CCRK Gene View. Provides basic gene information, including the number of products and poly(A) signals, and gene ontology terms for that gene. QGRS counts are displayed for the (30,2) and (45,3) categories together with non-overlapping versus overlapping QGRS. Additional QGRS information together with an exon/intron map is provided for each mRNA product. There are controls to navigate to any of the other views.
In the Gene View, the user has several options available to filter which QGRS will be displayed in the Data and Sequence Views. For example, the G-score, loop size, minimum size G-group and maximum QGRS length may be set by the user.
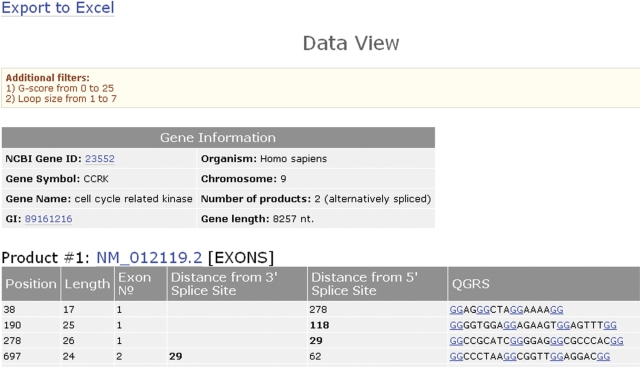
From the Gene View, the user may choose any of the other interfaces. The Data View displays the actual nucleotide sequence for each QGRS and locates its position in an exon, intron or near a poly(A) signal (Figure 3). The results of this page may be exported to Excel. There are two versions of the Data View. One view shows only non-overlapping QGRS, the other view displays all QGRS, overlapping or not.
Figure 3.
GRSDB2 CCRK Data View. A listing of the nucleotide sequences for all QGRS, the location of QGRS in exons, introns and near poly(A) signals. Results are shown for each product of the gene. What is shown is a truncated image of the view. The QGRS displayed satisfy the conditions that G-scores are in the range from 0 to 25 (which in effect restricts the output to QGRS with G-groups of size 2) and loop size is in the range from 1 to 7.
In the Sequence View, the nucleotide sequence for the entire gene is displayed. Exons are listed in purple and each QGRS is shown in yellow. The Graphic View gives the user a highly interactive visual tool to zoom in on any portion of the gene and analyze QGRS located in that section.
Structure and features of GRS_UTRdb
GRS_UTRdb contains information on the composition and distribution of QGRS in the UTRs of eukaryotic mRNA sequences.
Database users have 14 search fields to define queries. Fields such as GeneID, Gene Name/Symbol, GI and mRNA Accession Number allow the user to look for specific genes and mRNA products. There are fields for gene ontology function, process or component. The user may select which organism to search on. Also, ranges for the lengths of mRNA, 5′ UTR, 3′UTR or CDS may be specified.
Query results are summarized in a table from which the user may select a product for further analysis. GRS_UTRdb has six ways of viewing mRNA data: mRNA Map, Data View (no overlaps), Data View (with overlaps), Sequence View, Gene View, Alternate Products.
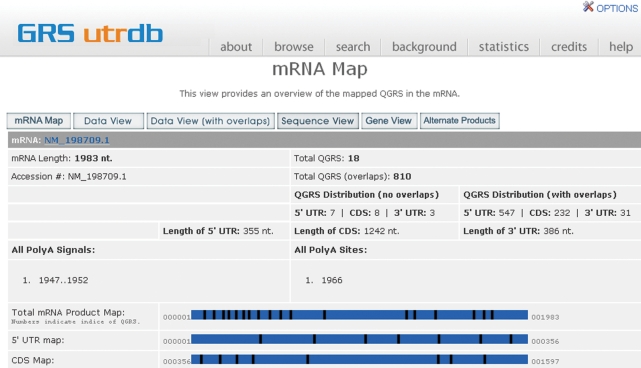
The mRNA map contains a table showing an overview of information about the product, including the lengths of the 5′ UTR, CDS and 3′ UTR (Figure 4). The number of QGRS found in each region is displayed. There is a visual map of the locations of QGRS within each region of the product.
Figure 4.
GRS_UTRdb NM_198709.1 mRNA (ARSB Gene) Map. Displays basic information about the 5′ UTR, CDS and 3′ UTR and the QGRS frequency in each region. A visual map of each region is displayed with the relative positions of QGRS depicted in the map.
The Data View shows the actual sequence of every QGRS in the product. The view gives the location of each QGRS in the 5′ UTR, CDS, 3′ UTR or near poly(A) signal. Also, G-score and the distance of QGRS from the nearest region boundaries are shown. There are two versions of the Data View, one that displays only non-overlapping QGRS and a view that shows all QGRS.
The sequence for the entire product may be found in the Sequence View and separate displays are shown for each region in the product. The QGRS are shown in a box, with each G-group in purple (Figure 5).
Figure 5.
GRS_UTRdb NM_198709.1 Sequence View. Shows the nucleotide sequence for the entire product, in separate displays for the 5′ UTR, CDS and 3′ UTR. QGRS are enclosed in a box with each G-group shown in purple.
The Gene View gives basic information about genes, including gene ontology terms and the location of poly(A) signals and sites. Each mRNA product is listed together with the distribution of QGRS there. The ‘Alternate Products’ tab takes the user directly to the QGRS information for each product associated with the gene.
CONCLUSIONS
GRSDB2 and GRS_UTRdb provide curated data on the composition and distribution of putative QGRS in the transcribed regions of a large number of alternatively processed eukaryotic genes. GRSDB2 is useful for studying G-quadruplexes near RNA-processing sites particularly those that are differentially processed. At present, it contains data for 29 288 genes encompassing 42 932 products from several eukaryotic organisms. More than 3 million QGRS have been mapped to these genes. The availability of large number of pre-mRNAs with mapped QGRS makes it possible to perform a variety of bioinformatics studies. The database website already offers a range of computational tools to aid large scale as well as individual gene analysis. The ‘Workbench’ can be used to perform computations on sets and sub-sets of genes in the database. The ‘Gene View’, ‘Data View’ and ‘Sequence View’ are useful for studying individual genes and their multiple products. The highly interactive ‘Graphic View’ is particularly useful for working with parts of the genes.
The new GRS_UTRdb offers a valuable resource for investigating G-quadruplexes in the UTRs of mRNA. Currently, it contains data for more than 16,000 eukaryotic mRNAs, including ∼27,000 QGRS which have been mapped to the 5′ UTRs. Like GRSDB2, GRS_UTRdb also displays QGRS data in a variety of modes with computational capabilities and links. At this point, it does not have a ‘Workbench’ facility. We are constantly adding new genes and new computational tools to the website. Since genes containing G-quadruplex motifs could be regulated through special mechanisms, one can expect for the gene function to correlate with G-quadruplex formation (32). We have classified the gene entries in our databases according to the gene ontology categories, which allows for queries with relevant terms.
Researchers interested in the functional relevance of G-quadruplex structure, in particular its role in regulating the gene expression at post-transcriptional level, will find both the databases to be of great value.
ACKNOWLEDGEMENTS
We thank Manuel Viotti for assistance with the testing and uploading data in the initial stages of this project. The authors would like to acknowledge Rumen Kostadinov, who developed the first version of GRSDB. We would also like to acknowledge Marcelo Halpern for technical assistance with the web server. This project was funded in part by a grant from the Provost Office of Ramapo College of New Jersey. Funding for the Open Access publication charges for this article was provided in part by the Divisions of Student Affairs and Academic Affairs of Ramapo College of New Jersey, and the Bergen County Academies, Hackensack, New Jersey.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halder K, Chowdhury S. Kinetic resolution of bimolecular hybridization versus intramolecular folding in nucleic acids by surface plasmon resonance: application to G-quadruplex/duplex competition in human c-myc promoter. Nucleic Acids Res. 2005;33:4466–4474. doi: 10.1093/nar/gki750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simonsson T. G-quadruplex DNA structures – variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 5.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 6.Kelland LR. Overcoming the immortality of tumour cells by telomere and telomerase based cancer therapeutics – current status and future prospects. Eur. J. Cancer. 2005;41:971–979. doi: 10.1016/j.ejca.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 7.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 9.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 10.Todd AK, Haider SM, Parkinson GN, Neidle S. Sequence occurrence and structural uniqueness of a G-quadruplex in the human c-kit promoter. Nucleic Acids Res. 2007;35:5799–5808. doi: 10.1093/nar/gkm609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wieland M, Hartig JS. RNA quadruplex-based modulation of gene expression. Chem. Biol. 2007;14:757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Sacca B, Lacroix L, Mergny JL. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimura Y, Martin CL, Vazquez-Lopez A, Spence SJ, Alvarez-Retuerto AI, Sigman M, Steindler C, Pellegrini S, Schanen NC, et al. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum. Mol. Genet. 2007;16:1682–1698. doi: 10.1093/hmg/ddm116. [DOI] [PubMed] [Google Scholar]
- 17.Bagga PS, Ford LP, Chen F, Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagga PS, Arhin GK, Wilusz J. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 1998;26:5343–5350. doi: 10.1093/nar/26.23.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arhin GK, Boots M, Bagga PS, Milcarek C, Wilusz J. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 2002;30:1842–1850. doi: 10.1093/nar/30.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalziel M, Nunes NM, Furger A. Two G-rich regulatory elements located adjacent to and 440 nucleotides downstream of the core poly(A) site of the intronless melanocortin receptor 1 gene are critical for efficient 3′ end processing. Mol Cell Biol. 2007;27:1568–1580. doi: 10.1128/MCB.01821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veraldi KL, Arhin GK, Martincic K, Chung-Ganster LH, Wilusz J, Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 2001;21:1228–1238. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruce SR, Dingle RW, Peterson ML. B-cell and plasma-cell splicing differences: a potential role in regulated immunoglobulin RNA processing. RNA. 2003;9:1264–1273. doi: 10.1261/rna.5820103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garneau D, Revil T, Fisette JF, Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang QS, Manche L, Xu RM, Krainer AR. hnRNP A1 associates with telomere ends and stimulates telomerase activity. RNA. 2006;12:1116–1128. doi: 10.1261/rna.58806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han K, Yeo G, An P, Burge CB, Grabowski PJ. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang E, Dimova N, Cambi F. PLP/DM20 ratio is regulated by hnRNPH and F and a novel G-rich enhancer in oligodendrocytes. Nucleic Acids Res. 2007;35:4164–4178. doi: 10.1093/nar/gkm387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 30.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kostadinov R, Malhotra N, Viotti M, Shine R, D’Antonio L, Bagga P. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonnal S, Schaeffer C, Creancier L, Clamens S, Moine H, Prats AC, Vagner S. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliver AW, Bogdarina I, Schroeder E, Taylor IA, Kneale GG. Preferential binding of fd gene 5 protein to tetraplex nucleic acid structures. J. Mol. Biol. 2000;301:575–584. doi: 10.1006/jmbi.2000.3991. [DOI] [PubMed] [Google Scholar]
- 36.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. reviews0004.1-reviews0004.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crnugelj M, Sket P, Plavec J. Small change in a G-rich sequence, a dramatic change in topology: new dimeric G-quadruplex folding motif with unique loop orientations. J. Am. Chem. Soc. 2003;125:7866–7871. doi: 10.1021/ja0348694. [DOI] [PubMed] [Google Scholar]
- 38.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 39.Risitano A, Fox KR. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kankia BI, Barany G, Musier-Forsyth K. Unfolding of DNA quadruplexes induced by HIV-1 nucleocapsid protein. Nucleic Acids Res. 2005;33:4395–4403. doi: 10.1093/nar/gki741. [DOI] [PMC free article] [PubMed] [Google Scholar]