Abstract
CATdb is a free resource available at http://urgv.evry.inra.fr/CATdb that provides public access to a large collection of transcriptome data for Arabidopsis thaliana produced by a single Complete Arabidopsis Transcriptome Micro Array (CATMA) platform. CATMA probes consist of gene-specific sequence tags (GSTs) of 150–500 bp. The v2 version of CATMA contains 24 576 GST probes representing most of the predicted A. thaliana genes, and 615 probes tiling the chloroplastic and mitochondrial genomes. Data in CATdb are entirely processed with the same standardized protocol, from microarray printing to data analyses. CATdb contains the results of 53 projects including 1724 hybridized samples distributed between 13 different organs, 49 different developmental conditions, 45 mutants and 63 environmental conditions. All the data contained in CATdb can be downloaded from the web site and subsets of data can be sorted out and displayed either by keywords, by experiments, genes or lists of genes up to 100. CATdb gives an easy access to the complete description of experiments with a picture of the experiment design.
INTRODUCTION
Transcriptome characterization by microarray technologies is a powerful tool for functional analysis of genes. The primary purpose of most of the experiments was finding candidate genes for further experimental work. Nevertheless, with the accumulation of data, a complementary usage of the transcriptome resource is the integration of large sets of data to infer, for instance, gene regulatory networks. Several databases dedicated to microarray data exist and can be distributed in three general classes (i) public repositories including ArrayExpress, Gene Expression Omnibus (GEO) and The Center for Information Biology Gene Expression Database (CIBEX) (1–3); (ii) general databases oriented toward tools for the analyses and displaying of different types of arrays, like Genevestigator or the Stanford Microarray Database (SMD) (4,5) and (iii) specific databases dedicated to a species like The Arabidopsis Information Resource, or the expression browser (eFP) from the Bio-Array Resource for Arabidopsis Functional Genomics, or specific to a life kingdom like the Plant Expression Database (PLEXdb) (6–8). Despite considerable and valuable efforts done to define and apply the Minimal Information About Microarray Experiment (MIAME) (9) recommendations, a recent survey of the data in public repositories indicated that data submission and quality are troublesome for integrating current microarray data (10). The diversity of transcriptome data and methods to analyse them is one of the problems for the occasional users. We have developed CATdb to manage the microarray data resource generated by the URGV transcriptome platform (http://www.versailles.inra.fr/urgv) and allow an easy access to the data by the community of biologists. We took advantage of the unique origin of the URGV-CATMA data to concentrate our effort on the quality of the data and to systematically collect a global view of each project with the details of the experiment design. Thus, CATdb provides an easy access to a large and growing set of microarrays named CATMA (Complete Arabidopsis Transcriptome Micro Array) (11). All the RNA samples are sent by collaborators at the URGV then checked for quality control, labelled and hybridized following normalized protocols. The scanning is performed with common settings and a unique normalization followed by a statistical analysis procedure is applied to each experiment as described subsequently.
CATMA MICROARRAYS
CATMA is a generic Arabidopsis thaliana microarray developed by a European consortium (12). The design of the probes for CATMA microarrays is different from the design of both the A. thaliana Agilent arrays (Palo Alto, CA, USA) and the ATH1 Affymetrix GeneChips (Santa Clara, CA, USA) (13) that use respectively oligo-nucleotide probes of 60 mers and sets of oligo-nucleotides of 25 mers. CATMA probes consist of gene-specific sequence tags (GSTs) of 150–500 bp that have been designed with SPADS (Specific Primers & Amplicons Design Software) (14). Tagged genes come from both the EuGene software prediction (15) and the annotation from The Institute for Genomic Research (TIGR).
The v2 version of CATMA contains 24 576 GST probes representing ∼85% of the predicted genes, 615 probes tiling the chloroplastic and mitochondrial genomes (v2.1) and 44 probes of non-protein coding genes (v2.2). A thorough benchmark study established the CATMA array as a mature alternative to the Affymetrix and Agilent platforms (16). The CATMA GSTs are also the basic materials in the AGRIKOLA (Arabidopsis Genomic RNAi Knock-out Line Analysis) European project focusing on the large-scale systematic RNAi silencing of Arabidopsis genes (http://www.agrikola.org/).
DATABASE CHARACTERISTICS AND CONTENTS
Primarily, CATdb was based on the schema and objects used in the ArrayExpress database (17). Then, the ArrayExpress schema has been adapted to our platform to include some new features. The main differences are: (i) the systematic addition of a figure describing the design of an experiment in standardized format, (ii) the possibility to manage a supplementary step with the pooling of samples or extracts and (iii) the storage of the statistical analyses using technical replicates (see the Data Analysis section).
The complete description of the experiments is submitted via a private web interface that helps to respect the MIAME instructions. CATdb generates the SOFT (Simple Omnibus Format in Text) format developed by the GEO repository (Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/). Data from the 1724 hybridizations in CATdb are also available either at GEO or at ArrayExpress. In the description of each project or experiment, there is the corresponding access number in GEO or ArrayExpress with a link to their respective web pages.
All the data submissions and analyses are performed in our laboratory, so the development of CATdb has been oriented by the visual approaches used by biologists at URGV, like using colours to encode the data values, to facilitate the analyses and comparisons of the data.
The increasing number of research projects involving CATMA microarrays shows that the CATMA arrays are an important tool for biologists. CATdb gives a public access to all the data produced by the URGV-CATMA platform even those that have not been published after one year. CATdb contains 46 projects with 1724 hybridizations corresponding to 627 different samples. The samples of these projects concern 13 types of organs: cells (73 samples), protoplasts (10), roots (116), hypocotyls (24), stems (18), leaves (129), flowers (36), pollen (2), siliques (4), seeds (17), whole aerial plants (43), plantlets (39) or whole plants (116). These samples are distributed between 49 different developmental conditions, 31 developmental stages, 45 mutants and 63 different abiotic/biotic stresses or treatments.
DATA ACCESS
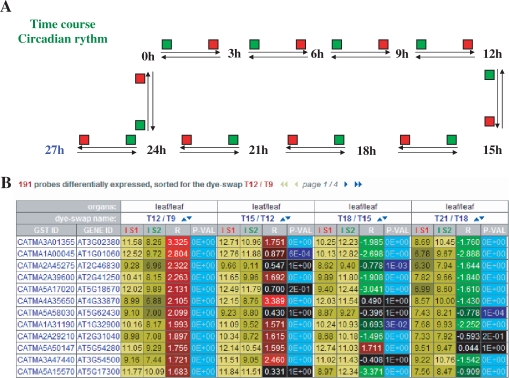
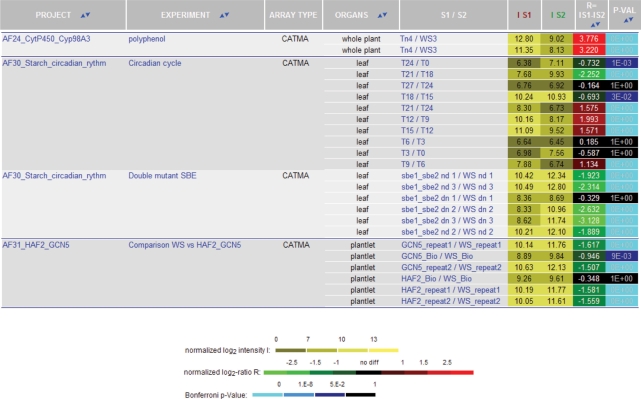
CATdb is a free web resource available at the following address: http://urgv.evry.inra.fr/CATdb. There are four different possibilities to select a subset of data. First, a list of all the available projects is displayed by default. A limited list may be obtained by querying the database by keywords. These keywords are searched for in both the description of the projects, i.e. coordinator name, experiment type, environmental or treatment factor, mutant name and the description of the samples, i.e. plant species, organs, treatments and type of arrays. Second, an experiment name may be selected in the project table giving access to the entire description of the corresponding experiment including a picture of the experiment design (Figure 1A). The swap column gives access to all the results of hybridizations organized by dye-swap for the selected experiment. Normalized log2 intensities, log2-ratios and Bonferroni P-values are given for each probe (Figure 1B). As this table is rather large, only the probes with statistically significant differential expression for a dye-swap are displayed on the screen. Nevertheless, the complete table may be downloaded as a tabulated text file. Third, from either a gene or a probe accession, one may obtain signal intensities and Bonferroni P-value for all the dye-swaps processed in all the projects (Figure 2). Furthermore, data may be sorted by project, organ or any statistics. For each probe, the associated features, i.e. sequence, quality of PCR results and if applicable, the tagged gene with functional annotation, are given. Fourth, from a list of genes or probe accessions, one obtains a table containing, for each selected probe, the log2-ratios for all the projects (Figure 3). The coloured display of the differential expression allows the comparison of the data for a list of up to 100 genes.
Figure 1.
Results of a query of CATdb for an experiment called ‘'C;ircadian cycle' and belonging to the project named ‘AF30_Starch_circadian_rythm’. The result includes (A) the experimental design that describes all the hybridizations with two colours, red and green, indicating the dye used for labelling each sample and (B) a table displaying for each dye-swap of the experiment and from left to right, the log2-intensities for samples 1 and 2, the log2-ratio and the Bonferroni P-value. In the example, results are sorted by the ratio values in the dye-swap between the leaf sample T12, extracted at 12 h after the start of the experiment, and the leaf sample T9, extracted at 9 h.
Figure 2.
Results of a query of CATdb with the name of a gene. The query was the gene AT1G32900. Data are displayed for all the projects in CATdb, but only the first four projects sorted by the Bonferroni P-values are shown here. From the left to the right, columns contain the project and the experiment names, the array type, the organ used, the sample name, the log2-intensities for both samples, the log2-ratio and the Bonferroni P-value.
Figure 3.
Results of a query of CATdb with a list of gene identifiers. From the left to the right, columns are the probe name, the corresponding gene name, the differential expression log2-ratio in all the different projects contained in CATdb. The differential expression is colour coded in green, red or black corresponding to the levels of log2-ratio in each swap analysed, and in grey for missing values. The correspondence between the colours and the log2-ratio values is given in a bar above the table. By rolling over a cell of the table, as shown by the arrows, one may display more information about either the projects, dye-swaps or expression values, depending on the selected cell.
All the public data contained in CATdb can be downloaded from an anonymous FTP (File Transfer Protocol) site (ftp://urgv.evry.inra.fr/CATdb). Users who have subscribed to the CATdb e-mailing list receive news about updates and new tools.
DATA ANALYSIS
Statistic methods were developed under the software R (R Development Core Team, http://www.R-project.org) in collaboration with the group ‘Statistics and Genome’ at UMR AgroParisTech/INRA MIA 518 and are available in the R package ‘Anapuce’ on their web site (http://www.inapg.fr/ens_rech/maths/outil_A.html). For each CATMA array, the raw data include the logarithm of median feature pixel intensity at wavelengths 635 nm (red) and 532 nm (green), no background is subtracted. A normalization per array is performed to remove systematic biases. First, spots that are considered badly formed features are excluded. Then, a global intensity-dependent normalization is performed using the lowess procedures (18) to correct the dye bias. Finally, for each block, the log-ratio median calculated over the values for the entire block is subtracted from each individual log-ratio value to correct effects on each block, as well as print-tip, washing and/or drying effects. At the end of the normalization step, a normalized log-ratio, which is equivalent to an expression difference (in log base 2) between the two samples co-hybridized on the same array, is given for each spot. It is equal to the raw log-ratio minus the lowess correction minus the block correction. A normalized logarithm intensity for each sample is also calculated. It is done according to the within-array correction proposed by Yang and Thorne (19), which is a redistribution of the correction calculated for the log2-ratio normalization on each channel.
To determine differentially expressed genes from a dye-swap, a paired t-test is performed on the log2-ratios. Since the number of observations per spot equals two, it is inadequate for calculating a specific variance. For this reason, it is assumed that the variance of the log2-ratios is the same for all spots. This solution has the main advantage to calculate an estimator over a large number of data, leading to a robust estimation of the variance and to a gain in the power of the test. Nevertheless, this solution should be applied with some precautions since some spots display an extreme specific variance (too small or too large) and prevent that the assumption of common variance is verified. Indeed spots with a too small specific variance decrease wrongly the estimate of the common variance and hence it could lead to increase the number of false positives, and spot with a too large variance increase wrongly the estimate of the common variance and hence it could lead to decrease the test power. For the above reasons, spots with extreme specific variance are excluded from the statistical analysis. The spots that are excluded are those with a ‘specific variance/common variance’ ratio smaller than the ‘alpha-quantile of a chi-squared distribution of one degree of liberty’ or greater than the ‘1-alpha-quantile of a chi-squared distribution of one degree of liberty’ with alpha equal to 0.0001. This rule stems from a direct application of Cochran's theorem. The raw P-values are adjusted by the Bonferroni method, which controls the Family Wise Error Rate (FWER) (20). When the Bonferroni P-value is lower than 0.05, the spot is declared differentially expressed. Spots with a missing P-value are spots with an extreme variance or genes for which one observation only is available. That is, when for one of the two arrays, the spot corresponding to the gene was a badly formed feature.
DATA QUALITY
Information on the CATMA probes and the corresponding genes is available in CATdb. This includes probe sequences and their estimated specificity, amplification efficiency, localization within genes (intron, exon) or between genes. All these annotations are graphically displayed in the genome database FLAGdb++ (21) and there are direct links from probes and genes in CATdb toward the probe loci in FLAGdb++. To validate transcriptome data, the biologist relies on quantitative RT-PCR applied to a set of genes exhibiting differential expression between two experimental situations. On the CATMA resource, quantitative RT-PCRs were done on more than 200 genes and CATMA results have been confirmed in more than 90% of the validations. The details of RT-PCR from tested genes are described in the publications associated to the different research projects using CATMA arrays. A list of these publications is available on the CATdb web site.
FUTURE PLANS
Based on the number of not yet public data, 4336 hybridized samples, stored in CATdb, the number of public projects is expected to double in the coming year. Updating data depends on the submission date of a project. As in most public repositories, data cannot be maintained under the private status more than one year and any data are publicly released after this period of time or before on the authors’ request.
CATMA is an ongoing project and new array designs will be released soon including 7189 new GSTs (collaboration with CATMA members) tagging the remaining annotated genes and different paralogues belonging to a gene family. Furthermore, probes for small RNA genes were designed by URGV in collaboration with O. Voinnet and L. Navarro (IBMP Strasbourg) and will be included in a future version. CATdb developments needed by the new designs are done in parallel.
ACKNOWLEDGEMENTS
The authors would like to thank Franck Samson for his help during the optimization of the code and Philippe Grevet for assistance and management of the ftp and web servers. Funding to pay the Open Access publication charges for this article was provided by INRA. The URGV CATMA resource has been funded by Génoplante.
Conflict of interest statement. None declared.
REFERENCES
- 1.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N, Lilja P, et al. ArrayExpress – a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, et al. NCBI GEO: mining tens of millions of expression profiles – database and tools update. Nucleic Acids Res. 2006;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikeo K, Ishi-I J, Tamura T, Gojobori T, Tateno Y. CIBEX: center for information biology gene expression database. C. R. Biol. 2003;326:1079–1082. doi: 10.1016/j.crvi.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann P, Hennig L, Gruissem W. Gene expression analysis and network discovery using Genevestigator. Trends Plant Sci. 2005;10:407–409. doi: 10.1016/j.tplants.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Demeter J, Beauheim C, Gollub J, Hernandez-Boussard T, Jin H, Maier D, Matese JC, Nitzberg M, Wymore F, et al. The Stanford MicroArray Datbase: implementation of new analysis and open source release of software. Nucleic Acids Res. 2007;35:D766–D770. doi: 10.1093/nar/gkl1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 8.Shen L, Gong J, Caldo RA, Nettleton D, Cook D, Wise RP, Dickerson JA. BarleyBase – An expression profiling database for plant genomics. Nucleic Acids Res. 2005;33:D614–D618. doi: 10.1093/nar/gki123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 10.Larsson O, Sandberg R. Lack of correct data format and comparability limits future integrative microarray research. Nat. Biotechnol. 2006;24:1322–1323. doi: 10.1038/nbt1106-1322. [DOI] [PubMed] [Google Scholar]
- 11.Crowe ML, Serizet C, Thareau V, Aubourg S, Rouzé P, Beynon JL, Hilson P, Weisbeek P, Van Hummelen P, et al. CATMA – A complete Arabidopsis GST database. Nucleic Acids Res. 2003;31:156–158. doi: 10.1093/nar/gkg071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao R, Bitton F, Caboche M, et al. Versatile gene-specific sequence tags for arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res. 2004;14:2176–2189. doi: 10.1101/gr.2544504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redman JC, Haas BJ, Tanimoto G, Town CD. Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J. 2004;38:545–561. doi: 10.1111/j.1365-313X.2004.02061.x. [DOI] [PubMed] [Google Scholar]
- 14.Thareau V, Déhais P, Serizet C, Hilson P, Rouzé P, Aubourg S. Automatic design of gene-specific sequence tags for genome-wide functional studies. Bioinformatics. 2003;19:2191–2198. doi: 10.1093/bioinformatics/btg286. [DOI] [PubMed] [Google Scholar]
- 15.Schiex T, Moisan A, Rouzé P. EuGene: an eukaryotic gene finder that combines several sources of evidence. Lect. Notes Comput. Sci. 2001;2066:111–125. [Google Scholar]
- 16.Allemeersch J, Durinck S, Vanderhaeghen R, Alard P, Maes R, Seeuws K, Bogaert T, Coddens K, Deschouwer K, et al. Benchmarking the CATMA microarray: a novel tool for Arabidopsis transcriptome analysis. Plant Physiol. 2005;137:588–601. doi: 10.1104/pp.104.051300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brazma A, Sarkans U, Robinson A, Vilo J, Vingron M, Hoheisel J, Fellenberg K. Microarray data representation, annotation and storage. Adv. Biochem. Eng. Biotechnol. 2002;77:113–139. doi: 10.1007/3-540-45713-5_7. [DOI] [PubMed] [Google Scholar]
- 18.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang YH, Thorne NP. Goldstein DR, editor. Normalization for two-color cDNA microarray data. Science and Statistics: A Festschrift for Terry Speed, IMS Lecture Notes–Monograph Series. 2003;Vol. 40:403–418. [Google Scholar]
- 20.Ge Y, Dudoit S, Speed TP. Resampling-based multiple testing for microarray data analysis. TEST. 2003;12:1–44. [Google Scholar]
- 21.Samson F, Brunaud V, Duchene S, De Oliveira Y, Caboche M, Lecharny A, Aubourg S. FLAGdb++: a database for the functional analysis of the Arabidopsis genome. Nucleic Acids Res. 2004;32:D347–D350. doi: 10.1093/nar/gkh134. [DOI] [PMC free article] [PubMed] [Google Scholar]