Abstract
G-protein coupled receptors (GPCRs) represent one of the most important families of drug targets in pharmaceutical development. GLIDA is a public GPCR-related Chemical Genomics database that is primarily focused on the integration of information between GPCRs and their ligands. It provides interaction data between GPCRs and their ligands, along with chemical information on the ligands, as well as biological information regarding GPCRs. These data are connected with each other in a relational database, allowing users in the field of Chemical Genomics research to easily retrieve such information from either biological or chemical starting points. GLIDA includes a variety of similarity search functions for the GPCRs and for their ligands. Thus, GLIDA can provide correlation maps linking the searched homologous GPCRs (or ligands) with their ligands (or GPCRs). By analyzing the correlation patterns between GPCRs and ligands, we can gain more detailed knowledge about their conserved molecular recognition patterns and improve drug design efforts by focusing on inferred candidates for GPCR-specific drugs. This article provides a summary of the GLIDA database and user facilities, and describes recent improvements to database design, data contents, ligand classification programs, similarity search options and graphical interfaces. GLIDA is publicly available at http://pharminfo.pharm.kyoto-u.ac.jp/services/glida/. We hope that it will prove very useful for Chemical Genomics research and GPCR-related drug discovery.
INTRODUCTION
The family of G-protein coupled receptors (GPCRs) represents one of the most important classes of pharmaceutical targets (1). Among the more than 1000 GPCRs encoded in the human genome, more than 400 are of potential therapeutic interest (2). Currently the drugs available on the market address only 30 GPCRs, which represent a small fraction of the GPCR target family. A large majority of human-derived GPCRs still remain promising drug targets, and thus a key goal of GPCR research related to drug design is to identify new ligands for such target GPCRs.
With the unprecedented accumulation of genomic information, databases and bioinformatics have become essential tools to guide GPCR research (3). The GPCRDB (2) and IUPHAR receptor database (IUPHAR-RD) (4) are representatives of widely used public databases covering GPCRs. These databases, which provide substantial data on the GPCR proteins and pharmacological information on receptor proteins containing GPCRs, are mainly focused on biological aspects of the GPCR gene products or proteins. In spite of the significance of ligand compounds as drug leads, the relationships between GPCRs and their ligands and/or chemical information on the ligands themselves are not yet fully covered.
On the other hand, there is increasing interest in publicly collecting and applying chemical as well as biological information in the post-genome era (5–8). This new trend is called ‘Chemical Genomics’, and it aims to identify all possible chemical ligands and drugs for all targets families (9,10). There is a vast amount of information on the interactions between small molecules and proteins/genes. However, compound–protein interactions have not yet been analyzed on a large scale, and there are no effective methods to extract meaningful information from the data in a comprehensive manner. Therefore, we need to integrate chemoinformatics and bioinformatics into a common computational platform for mining of Chemical Genomics data (11).
GLIDA (GPCR-Ligand DAtabase) is a public GPCR-related Chemical Genomics database designed to simultaneously mine biological information on GPCRs and chemical information on their ligands. It provides various analytical data regarding GPCR–ligand correlations by incorporating bioinformatics and chemoinformatics techniques, and thus it should prove very useful for GPCR-related drug discovery from the viewpoint of Chemical Genomics research. There have been several major improvements to GLIDA since it was last described in Ref. (12): (i) there are more increments in the entries of the ligands and the corresponding ligand–GPCR pairs; (ii) the ligands are originally classified using a new strategy; (iii) additional options are available within the similarity search program for the GPCRs and ligands and (iv) the graphical interface to display the correlation maps between GPCRs and ligands has been enhanced.
DATA CONTENTS
GLIDA contains three types of primary data: biological information on GPCRs, chemical information on their ligands and information on binding of the GPCR–ligand pairs. The GPCR entries were acquired from human, mouse and rat entries deposited in the GPCRDB because these three species include sufficient information regarding ligands, and rats and mice are representative model animals used in drug discovery research. The ligand-binding information was manually collected and curated using various public web sites and commercial databases such as the IUPHAR-RD, PubMed (5), PubChem (5), DrugBank (13), Ki Database (14) and MDL ISIS/Base 2.5. Table 1 indicates the size and scope of the GLIDA database. In particular, we have dramatically expanded the entry number of ligands and the corresponding ligand–GPCR pairs. The latest GLIDA version includes 24 077 ligand entries and 39 140 GPCR–ligand pair entries, representing nearly 35-fold and 20-fold increases, respectively, since the last publication of GLIDA in 2006. The total number of GPCR entries remains unchanged, but entries with associated ligand information have increased slightly, suggesting that it is difficult to de-orphan the GPCRs whose ligands have not yet been identified (15).
Table 1.
The current numbers of GLIDA ligands and GPCRs and their respective links.
| Information item | Number of entries |
|---|---|
| GPCR entries | 3738 |
| Links to Entrez Gene | 3073 |
| Links to GPCRDB | 3738 |
| Links to UniProt | 3738 |
| Links to IUPHAR | 446 |
| Links to KEGG | 595 |
| Ligand entries | 24 077 |
| Cas registry number | 2425 |
| Molecular structure | 23 216a |
| Links to PubChem | 1821 |
| Links to ChEBI | 103 |
| Links to KEGG | 664 |
| Links to DrugBank | 479 |
| Cluster number | 300b |
| GPCR–ligand pair entries | 39 140 |
| GPCR entries | 410 |
| Ligand entries | 24 077 |
| Activity | |
| Agonist | 8305 |
| Full Agonist | 2325 |
| Partial Agonist | 262 |
| Antagonist | 28 132 |
| Inverse Agonist | 116 |
aMolecular structures consist of MDL MOL files and original files converted into KEGG atom types. The numbers of MDL MOL files and KEGG-type files are 23 216 and 23 214, respectively. PCA calculation was performed for 23 214 KEGG-type files.
bThis cluster number (300) is different from the number of the selected principal components (314). No compounds were assigned to 14 principal components.
GPCR and ligand data
The database lists general information on GPCR and ligand data, respectively. The general information table listing GPCRs contains gene names, family names, protein sequences (in fasta format) and links to other biological databases, such as GPCRDB, UniProt (16), IUPHAR-RD, Entrez Gene (17) and KEGG (18). The ligand result page provides a general information table containing names, molecular structures, CAS registry numbers, formulas, molecular weights, structure files and links to PubChem, KEGG, ChEBI (8) and DrugBank that are in publicly available chemical databases.
Information on binding of GPCR–ligand pairs
The interaction information relating GPCRs to particular ligands, a key issue for GPCR-related drug discovery, is deposited in a relational database. GLIDA allows users to retrieve GPCR–ligand-binding information dynamically and continuously. When users retrieve a GPCR (or ligand) entry, its result page displays all entries showing the corresponding ligands (or GPCR) entries with their binding activity types, as well as references. The references are hyperlinked with the corresponding PubMed literature. The activity types include agonist, antagonist and full, partial or inverse agonist (Table 1). Here the detail annotations such as full, partial or inverse agonist are not finished yet. The ligands classified as agonists are possible full agonists or partial agonists. Inverse agonists can be also contained among the antagonists.
WEB INTERFACE AND APPLICATION
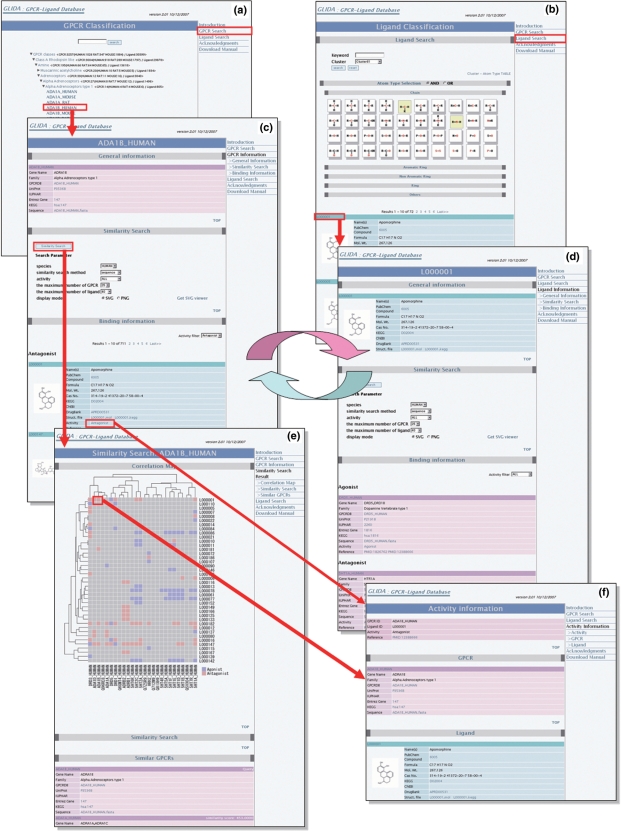
GLIDA is available at http://pharminfo.pharm.kyoto-u.ac.jp/services/glida/. The web interface of GLIDA includes a GPCR search page (Figure 1a) and a ligand search page (Figure 1b). Each page consists of a classification menu and a keyword search box. The users can search a GPCR (or ligand) manually using the classification tool, or automatically by using the keyword search function. Every GPCR (or ligand) has its own results page (Figure 1c or d) containing a general information table regarding a GPCR (or ligand), a table of its correlated ligands (or GPCRs) and a menu button to carry out a similarity search and correlation analysis.
Figure 1.
A screenshot of GLIDA showing linked relations among search pages (a and b), result pages (c and d), an analytical report page (e), and a binding information page (f). The analytical report page consists of a correlation map and a list resulting from a similarity search. Red and blue colors of the spots on the correlation map indicate the ligand activities of antagonists including inverse agonist and agonists including full/partial agonist, respectively.
Classification of GPCRs and ligands
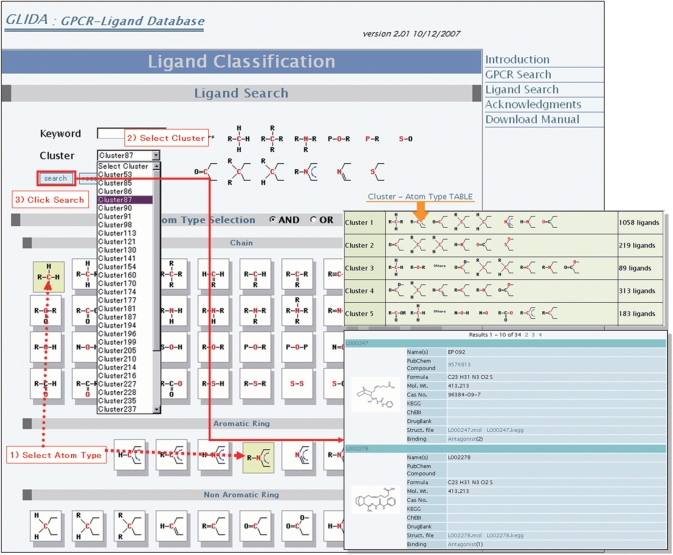
The GPCR classification table on the search page was adapted from the phylogenetic tree within the GPCRDB information system (http://www.gpcr.org/7tm/phylo/phylo.html). The GPCR classification table displays the entries of the corresponding GPCRs at the tree branches, and these are hyperlinked with the corresponding result pages (Figure 1a). GLIDA also provides an original ligand classification (Figures 1b and 2). With the great increase in ligand entries, we have to improve our method of classifying all the ligands in GLIDA. Hierarchical clustering and its tree representation, which were used in the old version of GLIDA, are unsuitable for the data mining of huge chemical databases. We therefore have adopted principal component analysis (PCA) for clustering of 23 214 ligand structures in this new version, as follows. We generated frequency profiles of the atoms and the bonds converted into the KEGG atom types from MDL MOL files of ligand entries (19). The KEGG-type profile for each ligand is shown in ‘Struct. file’ item of general information table of GLIDA. PCA was applied to the data matrix consisting of 700 KEGG-type features’ columns and 23 214 ligand entries’ rows. The resulting principal components (PCs) constitute a new set of linearly independent, orthogonal axes that capture the directions of maximum variance in the data. The samples (chemical compounds) were then projected onto these PC axes. Herein, we used the top 314 PCs as seeds of clusters that account for >80% (cumulative proportion) of the total variance. Finally, each compound was assigned to the PC cluster having the maximum score among the 314 PCs. In order to annotate the features of each cluster (PC), we selected for each PC the atom types and their bonds corresponding to the top 10 loadings having the largest magnitude. The ligand classification page displays a table of all the atom types selected by PCA (Figure 2). By clicking on some of the atoms in this table, users can search clusters that include the selected atom types. Consequently, the ligands relevant to users’ interests are included in the retrieved cluster.
Figure 2.
A screenshot of the ligand search process on the ligand classification page. Users can search the ligands from two starting points: keyword search and cluster selection. If they have a chemical structure of their query compound, the ligand search is performed using the cluster selection tool as follows. Selecting a set of atom types (step 1) that the query compound contains, the pull-down menu of cluster selection displays the list of the only clusters that include selected atom types as the principal components (step 2). By selecting a cluster from the list, users can check the principal component's atoms on the upper right section of the page. Finally, upon clicking the search button, GLIDA displays the list of all ligands classified in the selected cluster (step 3). The ‘Atom Type TABLE’ button links the user to the page showing the cluster size and representative atom types for each cluster.
Similarity search and correlation map for GPCRs and ligands
The fact that similar proteins bind similar ligands is the underlying principle of the Chemical Genomics approach to drug discovery (11). GLIDA has a variety of similarity search functions for GPCRs and ligands, respectively, on its result pages (Figure 1c or d). Alignment scores for protein sequences generated by the BLAST algorithm provide similarity measures for GPCRs. In addition to sequence similarity, gene expression patterns in tissue origins and developmental stages were used as similarity measures. The expression data for each GPCR was generated from the EST sequences in different libraries served from NCBI/Unigene (http://www.ncbi.nlm.nih.gov/UniGene/ddd.cgi). We can thereby retrieve the GPCRs that present tissue-/stage-specific distribution similar to a query GPCR. For example, co-expression information on specific GPCRs enables us to speculate about GPCR-heterodimerization that might have an effect on their activity (1). Ligand similarity is defined by the dissimilarity (distance) of frequency profile patterns generated from the constitutive atoms and bonds of the chemical structure, using the KEGG atom types (19,20). From the similarity search, the most similar GPCRs (or ligands) within the users’ selected parameters are retrieved and listed with their similarity scores on an analytical report page (Figure 1e). In the latest GLIDA version, various parameters have been added as search options, such as selections of species, ligand activities, displayed number of GPCRs/ligands and map graphical mode. As another result of similarity search calculations, GLIDA illustrates the correlation map (Figure 1e) showing homologous GPCRs (or ligands) and their ligands (or GPCRs) that are retrieved. This map shows spots that match the GPCRs and their ligands in a 2D matrix. The ordering along the x-axis and the y-axis are calculated respectively by two-way clustering of the GPCRs and the ligands, based on their similarities. In particular, the ordering along the x- and y-axes allows users to evaluate the sequence similarities among GPCRs and the correlation coefficients among ligands simultaneously. By analyzing the correlation patterns between GPCRs and ligands that are illustrated by these maps, we can gain detailed knowledge about their interactions. We can then utilize this information to infer possible candidates for development of GPCR-specific drugs. Furthermore, we have enhanced a graphical interface to display the correlation map between GPCRs and ligands. Graphics are an important tool to aid visualization and interpretation of high-dimensional data. The old version of GLIDA used only the PNG (Portable Network Graphics) format to display a GPCR–ligand correlation map. Due to the great increase in entries, the latest GLIDA version introduces the SVG (Scalable Vector Graphics) format, which is adaptable to an enormous correlation map size. The SVG vector image can be scaled indefinitely without loss of image quality, while the PNG bitmap image cannot. Users must install the free plug-in software on their computer in advance to use the SVG format (http://www.adobe.com/svg/viewer/install/). In the case of uninstalled devices, PNG representation should be selected as a graphical mode. Figure 1 shows an example of the GPCR–ligand search and analysis process starting from a GPCR query using GLIDA.
DISCUSSION AND FUTURE DIRECTIONS
GLIDA provides a unique database useful for GPCR-related Chemical Genomics research and drug discovery. GLIDA is distinct from other public Chemical Genomics databases because it contains original, GPCR-specific chemical entries and offers a common mining platform of bioinformatics and chemoinformatics. GLIDA provides several advantages over other databases, in that a search can be started either from a GPCR or from a ligand. Thus, searches can be carried out in a dynamic and user-friendly way. GLIDA's coverage of chemical and biological information simultaneously also provides an advantage to users by saving them the time and labor required to search multiple databases. The ligand search page is another distinct characteristic of GLIDA, in that it displays the structural distribution of ligands. It thereby facilitates research on GPCR-related drugs by incorporating structural aspects of the ligand compounds into the search. The analytical report pages resulting from the calculated structural similarities of GPCRs and ligands can give the user deep insights into the GPCR–ligand relationships. The lists of neighboring ligands (or GPCRs) and the correlation maps are useful visualization tools for analyzing correlations among the structural features and the GPCR–ligand-binding properties. Because this database system can be applied to proteins other than the GPCR family, it may also be considered as a promising database for other types of Chemical Genomics research. One critical issue is how to define similarity metrics for proteins and ligands, because the underlying principle of GLIDA is that similar receptors bind similar ligands. For example, ligand similarity can be defined by manifold representations such as graph, fingerprint and descriptors. Protein similarity can be also measured in different ways such as overall sequence homology (phylogenetic relationships), consensus motifs, common binding sites, 3D structures and reported functional annotations. Therefore we will add new menus incorporating these various similarity metrics for GPCRs and ligands. GLIDA will be updated continuously. In particular, we are now planning to add the drawing tool of chemical structures and to expand the ligand-searching function for an arbitrary chemical query.
ACKNOWLEDGEMENTS
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, from the JSPS, KAKENHI, Grant-in-Aid for Publication of Scientific Research Results and from the Ministry of Health, Labour and Welfare of Japan. Financial support from the SUNTORY INSTITUTE FOR BIOORGANIC RESEARCH, the TATEISI SCIENCE AND TECHNOLOGY FOUNDATION and the Okawa Foundation for Information and Telecommunications is gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.George SR, O’Dowd BF, Lee SP. G-protein-coupled receptor oligomerization and its potential for drug discovery. Nature Rev. Drug Discov. 2002;1:808–820. doi: 10.1038/nrd913. [DOI] [PubMed] [Google Scholar]
- 2.Horn F, Bettler E, Oliveira L, Campagne F, Cohen FE, Vriend G. GPCRDB information system for G protein-coupled receptors. Nucleic Acids Res. 2003;31:294–297. doi: 10.1093/nar/gkg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strachan R, Ferrara G, Roth BL. Screening the receptorome: an efficient approach for drug discovery and target validation. Drug Discov. Today. 2006;11:708–716. doi: 10.1016/j.drudis.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Foord SM, Bonner TI, Neubig RR, Rosser EM, Pin JP, Davenport AP, Spedding M, Harmar AJ. International Union of Pharmacology. XLVI. G Protein-Coupled Receptor List. Pharmacol. Rev. 2005;57:279–288. doi: 10.1124/pr.57.2.5. [DOI] [PubMed] [Google Scholar]
- 5.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiber SL. Stuart Schreiber: biology from a chemist's perspective. Interview by Joanna Owens. Drug Discov. Today. 2004;9:299–303. doi: 10.1016/S1359-6446(04)03063-6. [DOI] [PubMed] [Google Scholar]
- 7.Goto S, Okuno Y, Hattori M, Nishioka T, Kanehisa M. LIGAND: database of chemical compounds and reactions in biological pathways. Nucleic Acids Res. 2002;30:D402–D404. doi: 10.1093/nar/30.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooksbank C, Cameron G, Thornton J. The European Bioinformatics Institute's data resources: towards systems biology. Nucleic Acids Res. 2005;33:D46–D53. doi: 10.1093/nar/gki026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zerhouni E. The NIH Roadmap. Science. 2003;302:63–72. doi: 10.1126/science.1091867. [DOI] [PubMed] [Google Scholar]
- 10.Dobson CM. Chemical space and biology. Nature. 2004;432:824–828. doi: 10.1038/nature03192. [DOI] [PubMed] [Google Scholar]
- 11.Klabunde T. Chemogenomic approaches to drug discovery: similar receptors bind similar ligands. Br. J. Pharmacol. 2007;152:5–7. doi: 10.1038/sj.bjp.0707308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okuno Y, Yang J, Taneishi K, Yabuuchi H, Tsujimoto G. GLIDA: GPCR-ligand database for chemical genomic drug discovery. Nucleic Acids Res. 2006;34:D673–D677. doi: 10.1093/nar/gkj028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roth BL, Lopez E, Beischel S, Westkaemper RB, Evans JM. Screening the receptorome to discover the molecular targets for plant-derived psychoactive compounds: a novel approach for CNS drug discovery. Pharmacol. Ther. 2004;102:99–110. doi: 10.1016/j.pharmthera.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Civelli O. GPCR deorphanizations: the novel, the known and the unexpected transmitters. Trends Pharmacol. Sci. 2005;26:15–19. doi: 10.1016/j.tips.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 16.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Research. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hattori M, Okuno Y, Goto S, Kanehisa M. Development of a Chemical Structure Comparison Method for Integrated Analysis of Chemical and Genomic Information in the Metabolic Pathways. J. Am. Chem. Soc. 2003;125:11853–11865. doi: 10.1021/ja036030u. [DOI] [PubMed] [Google Scholar]
- 20.Kotera M, Okuno Y, Hattori M, Goto S, Kanehisa M. Computational assignment of the EC numbers for genomic-scale analysis of enzymatic reactions. J. Am. Chem. Soc. 2004;126:16487–16498. doi: 10.1021/ja0466457. [DOI] [PubMed] [Google Scholar]