Abstract
The genome sequences of a large number of metazoan species are now known. As multiple closely related genomes are sequenced, comparative studies that previously focussed on only pairs of genomes can now be extended over whole clades. The orthologous and paralogous transcripts in clades (OPTIC) database currently provides sets of gene predictions and orthology assignments for three clades: (i) amniotes, including human, dog, mouse, opossum, platypus and chicken (17 443 orthologous groups); (ii) a Drosophila clade of 12 species (12 889 orthologous groups) and (iii) a nematode clade of four species (13 626 orthologous groups). Gene predictions, multiple alignments and phylogenetic trees are freely available to browse and download from http://genserv.anat.ox.ac.uk/clades. Further genomes and clades will be added in the future.
INTRODUCTION
New technologies and reduced costs are driving a marked increase in the numbers of genomes that are being sequenced. This steep rise in data presents opportunities for predicting evolutionary relationships of genes not between pairs of genomes, as previously, but instead among genomes from a clade of closely related species. Computational tools for gene prediction, orthology assignment and multiple alignment are now needing to be developed using phylogenetic approaches. To meet this challenge, we have developed a pipeline for gene prediction and orthology assignment for any clade of genomes (Heger and Ponting, in press). The current release of the orthologous and paralogous transcripts in clades (OPTIC) database contains three clades: 12 species from the genus Drosophila, an amniotic clade of five mammals with chicken as outgroup, and four Caenorhabditis nematodes (Table 1).
Table 1.
Gene sets and orthology assignments in three clades
| Species | Genes | Genes with orthologs (%) | Orphaned genes (%) |
|---|---|---|---|
| D. melanogaster* | 13 836 | 13 563 (98) | 273 (2) |
| D. simulans | 13 203 | 12 318 (93) | 885 (7) |
| D. sechellia | 15 467 | 14 356 (93) | 1111 (7) |
| D. erecta | 14 199 | 13 471 (95) | 728 (5) |
| D. yakuba | 14 971 | 14 218 (95) | 753 (5) |
| D. ananassae | 14 337 | 13 205 (92) | 1132 (8) |
| D. pseudoobsura | 12 304 | 11 609 (94) | 695 (6) |
| D. persimilis | 12 973 | 11 876 (92) | 1097 (8) |
| D. willistoni | 13 144 | 11 360 (86) | 1784 (14) |
| D. virilis | 12 017 | 11 096 (92) | 921 (8) |
| D. mojavensis | 11 717 | 10 883 (93) | 834 (7) |
| D. grimshawi | 11 800 | 11 011 (93) | 789 (7) |
| C. elegans* | 20 093 | 14 037 (70) | 6056 (30) |
| C. remanei | 18 137 | 14 961 (82) | 3176 (18) |
| C.PB2801 | 21 931 | 17 759 (81) | 4172 (19) |
| C. briggsae | 18 388 | 13 460 (73) | 4928 (27) |
| H. sapiens* | 22 611 | 19 339 (86) | 3272 (14) |
| M. musculus* | 24 442 | 20 758 (85) | 3684 (15) |
| C. familiaris* | 19 314 | 18 066 (94) | 1248 (6) |
| M. domestica* | 19 597 | 18 123 (92) | 1474 (8) |
| O. anatinus* | 18 596 | 15 312 (82) | 3284 (18) |
| G. gallus* | 16 715 | 13 893 (83) | 2822 (17) |
Gene sets marked with an asterisk (*) were obtained from ENSEMBL, whereas all others have been predicted by the pipeline. Orphans represent genes that have no ortholog in any of the other genomes in the clade. These will represent results of heuristic failures in our ortholog prediction pipeline or in gene predictions, as well as true gene losses.
The pipeline predicts orthology for both orthologous groups and simple 1:1 ortholog sets. Here, orthologous groups contain orthologs and in-paralogs but exclude out-paralogs (1), those duplicated genes that were each present in the last common ancestor of a clade. Simple 1:1 ortholog sets are derived from orthologous groups by examining the gene tree and extracting sub-trees that contain exactly one gene per species. To enable inferences of gene duplication or loss, or positive selection on individual codons, we supply amino acid or nucleotide multiple sequence alignments, and phylogenetic trees, for each orthologous group. All data may be searched or downloaded freely from http://genserv.anat.ox.ac.uk/download/clades.
Database construction
The pipeline requires a set of genome sequences and ENSEMBL (2) gene sets for each genome. If a gene set for a genome is unavailable, we predict transcripts by homology from a reference transcript set and thereafter automatically derive a gene set from them (Heger and Ponting, in press). A quality control step removes partial predictions and marks those predictions as pseudogenes that contain in-frame stop-codons and frameshift insertions and deletions. Both genes and pseudogenes comprise a predicted gene set. ENSEMBL and predicted gene sets are then submitted to an orthology assignment process. A full description of the pipeline, including parameter settings, is provided on the web site. Briefly, the pipeline implements the following steps:
Gene prediction by homology from a transcript set using Exonerate (3).
Graph-based grouping of genes from all species into clusters.
Multiple alignment of translated exons using MUSCLE (6).
Estimation of phylogenetic tree topology using NJTree (7).
Decomposition of clusters into orthologous groups.
Branch length estimation using codeml from the PAML package (8).
Computation of simple 1:1 ortholog sets.
Data are stored in a relational database and gene predictions are displayed within a GMOD genome browser (http://www.gmod.org). Software is open source and available without charge on request to the authors.
Database contents
For the current release, we have applied our pipeline to three metazoan clades (Table 1) each containing between 4 and 12 species. Genes were predicted for Drosophila and Caenorhabditis species’ genome assemblies using D. melanogaster (9) and C. elegans (10) protein-coding transcripts as templates. Mammalian and chicken gene sets were from ENSEMBL release 42 (2). The web server provides an up-to-date list of genome assemblies for the current release.
We find 12 889 orthologous groups in the Drosophila clade, 17 443 groups in the amniotic clade and 13 626 groups in the four Caenorhabditis species. Of these, 10 563 orthologous groups in the Drosophila clade, 9675 groups in the amniotic clade and 6545 groups in the Caenorhabditis clade contain the full species complements. The numbers of simple 1:1 ortholog sets are smaller (5241, 7587, and 5987 for the three clades, respectively) owing to gene duplications and absences from incomplete assemblies.
For each orthologous group, we provide:
Transcript predictions: Predicted transcripts are available as exonic genomic coordinates, and as peptide and coding sequences.
Orthologs: Orthologous groups and simple 1:1 ortholog sets.
Multiple alignments: Multiple alignments of transcripts and genes within an orthologous group are provided both as aligned nucleic acid sequences and as aligned peptide sequences. Frameshift insertions or deletions in pseudogenes have been removed, and stop-codons have been masked in order to facilitate downstream analyses. Genes have been aligned by concatenating exons of all transcripts while maintaining frame.
Phylogenetic trees: For each orthologous group, we provide a phylogenetic tree. The topology of the tree has been calculated from NJTree, while branch lengths (nucleotide substitutions per site) have been assigned using PAML.
Database access and web service
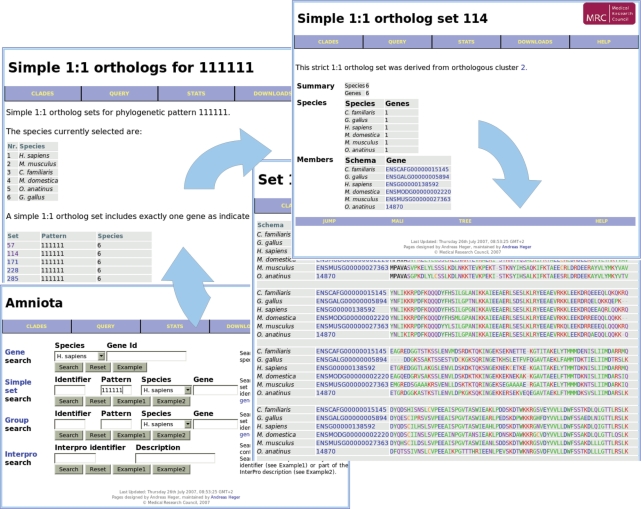
The web service permits interactive data querying and browsing of orthologous groups and simple 1:1 ortholog sets for each clade (Figure 1). Species distributions of orthologous groups are denoted by phylogenetic profiles denoting the presence (‘+’) or absence (‘0’) of one or more genes in a group. For example, a search for orthologous groups in the amniotic clade with the phylogenetic profile ‘+ + +000’ lists 542 orthologous groups that contain genes in human, mouse and dog, but have no orthologs in opossum, platypus and chicken.
Figure 1.
Browsing the orthology database. A sample session starts with a query for all simple 1:1 ortholog sets (bottom left). It continues with a list of all simple 1:1 ortholog sets containing all six species from the amniotic clade, then by a selection of one particular ortholog set (number 114), and finally with a viewing of the gene-based multiple sequence alignment.
In queries for simple 1:1 ortholog sets, ‘1’ indicates that exactly one copy of this gene is present and ‘–’ indicates that this particular species should not be considered. Thus, the profile ‘111–––’ applied to simple 1:1 ortholog sets yields 13 788 simple 1:1 ortholog sets that contain exactly one gene in human, dog and mouse, and any number of homologs in opossum, platypus or chicken.
For each orthologous group and simple 1:1 ortholog set, multiple alignments and a phylogenetic tree may be displayed. A synteny viewer also allows an assessment of whether orthologs occur in regions of conserved synteny. Genes of particular interest can be located either by identifier or by genomic location. Computational biologists interested in performing large-scale analyses can download complete datasets from the download area.
OUTLOOK
OPTIC is designed to provide precalculated phylogenetic datasets that are of benefit to clade genomic analyses. Our approach complements other existing projects (2,7,12, 12) in four respects: (i) we apply the pipeline to diverse, and not just experimental model, organisms; (ii) we define clades with respect to phylogenetic distances that are amenable to evolutionary rate analysis (roughly, where the number of synonymous substitutions per synonymous site is <2.0 (5)); (iii) our orthology relationships are inferred by considering all species equally, in a phylogenetic approach and (4) we use all exons across all alternative transcripts as opposed to the longest transcripts only. A particularly useful feature of OPTIC is its provision of multiple alignments either for genes as concatenated exons, or for alternative transcripts.
We plan to update gene predictions and orthology assignments and add more genomes and clades when they become available.
ACKNOWLEDGEMENTS
This study was funded by Medical Research Council, UK. We are grateful to Leo Goodstadt for many helpful discussions. We would like to thank the various genome sequencing centers and ENSEMBL for making their genomic data and gene sets freely available for download. Funding to pay the Open Access publication charges for this article was provided by Medical Research Council, UK.
Conflict of interest statement. None declared.
REFERENCES
- 1.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 2.Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodstadt L, Ponting CP. Phylogenetic reconstruction of orthology, paralogy, and conserved synteny for dog and human. PLoS Comput. Biol. 2006;2:e133. doi: 10.1371/journal.pcbi.0020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar RC. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Coghlan A, Ruan J, Coin LJ, Heriche J, Osmotherly L, Li R, Liu T, Zhang Z, et al. Treefam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z. Paml: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 9.Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM. Flybase: genomes by the dozen. Nucleic Acids Res. 2007;35:D486–491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bieri T, Blasiar D, Ozersky P, Antoshechkin I, Bastiani C, Canaran P, Chan J, Chen N, Chen WJ, et al. Wormbase: new content and better access. Nucleic Acids Res. 2007;35:D506–D510. doi: 10.1093/nar/gkl818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Brien KP, Remm M, Sonnhammer ELL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huerta-Cepas J, Dopazo H, Dopazo J, Gabaldon T. The human phylome. Genome Biol. 2007;8:R109. doi: 10.1186/gb-2007-8-6-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heger A, Ponting CP. Evolutionary rate analysis of orthologues and paralogues from twelve Drosophila genomes. Genome Res. 2007:in press. doi: 10.1101/gr.6249707. [DOI] [PMC free article] [PubMed] [Google Scholar]