Abstract
miRBase is the central online repository for microRNA (miRNA) nomenclature, sequence data, annotation and target prediction. The current release (10.0) contains 5071 miRNA loci from 58 species, expressing 5922 distinct mature miRNA sequences: a growth of over 2000 sequences in the past 2 years. miRBase provides a range of data to facilitate studies of miRNA genomics: all miRNAs are mapped to their genomic coordinates. Clusters of miRNA sequences in the genome are highlighted, and can be defined and retrieved with any inter-miRNA distance. The overlap of miRNA sequences with annotated transcripts, both protein- and non-coding, are described. Finally, graphical views of the locations of a wide range of genomic features in model organisms allow for the first time the prediction of the likely boundaries of many miRNA primary transcripts. miRBase is available at http://microrna.sanger.ac.uk/.
INTRODUCTION
MicroRNAs (miRNAs) are short RNA sequences expressed from longer transcripts encoded in animal, plant and virus genomes, and recently discovered in a single-celled eukaryote (1,2). miRNAs regulate the expression of target genes by binding to complementary sites in their transcripts to cause translational repression or transcript degradation (3). Translational repression is thought to be the primary mechanism for imperfect target duplexes in animals, with transcript degradation the dominant mechanism for largely perfect matches found throughout plant target transcripts. miRNAs have been implicated in processes and pathways such as development, cell proliferation, apoptosis, metabolism and morphogenesis, and in diseases including cancer (4,5).
miRBase is the primary repository and database resource for miRNA data. The database has three main functions:
miRBase::Registry provides a confidential service for the independent assignment of names to novel miRNA genes prior to their publication in peer-reviewed journals. Over 70 publications describing novel miRNA genes have made use of this service, and registration is a requirement of many journals.
miRBase::Sequences provides miRNA sequence data, annotation, references and links to other resources for all published miRNAs. The database (release 10.0) contains over 5000 sequences from 58 species.
miRBase::Targets provides an automated pipeline for the prediction of targets for all published animal miRNAs. The current release of the database (v5) predicts targets in over 500 000 transcripts for all miRNAs in 24 species. The target prediction pipeline and algorithms have been described elsewhere (6,7).
The miRNA nomenclature scheme has been presented and discussed previously (6,8,9). Novel miRNAs require cloning or expression evidence, and should be submitted only after a manuscript describing their identification is accepted for publication. Assigned names should then be incorporated into the final version of the manuscript prior to publication. Obvious homologues of miRNAs validated in closely related species need not be experimentally verified and may be submitted at any time. Primary features of the nomenclature scheme are:
The miRNA name contains a three or four letter species prefix and a numeric suffix (e.g. hsa-mir-212).
A mature miRNA sequence may be predicted to be expressed from more than one hairpin precursor locus, denoted with further numeric suffixes (e.g. dme-mir-6-1 and dme-mir-6-2).
Related hairpin loci expressing related mature miRNA sequences have lettered suffixes (e.g. mmu-mir-181a and mmu-mir-181b).
Plant miRNA genes are given names of the form ath-MIR166a. Lettered suffixes describe distinct loci expressing all related mature miRNAs; numeric suffixes are not used.
Viral miRNA names conventionally relate to the locus from which the miRNA derives (e.g. ebv-mir-BART1 from the Epstein Barr virus BART locus).
However, it is important to note that a short name cannot always encode complex information such as orthology and paralogy relationships. In some cases, the short name is a pragmatic choice that is the most consistent of conflicting representations of these sequence relationships. While the names provide a guide of family and function, they should not therefore be relied upon to confer any complex meaning. Instead, dedicated fields in the database provide information about gene and mature miRNA sequence families.
The published miRNA literature is huge. Readers are referred to a number of comprehensive reviews of miRNA structure, biogenesis and function (4,10–12). Here, we focus on specific issues and points of interest with respect to the provision of miRNA data in the miRBase database.
miRBase DATA AND UPDATES
How many miRNA genes?
The number of miRNA hairpin loci in the miRBase database continues to grow rapidly, from 2909 in 36 genomes (June 2005, release 7.0) to 5071 in 58 genomes (August 2007, release 10.0) in the past 2 years. The number of miRNAs in a genome has been the subject of much discussion in the literature. Early estimates of the number of miRNAs in the worm and human genomes were put at 123 and 255, respectively (13,14). However, these estimates were based largely on conservation studies. It is now clear that many miRNAs may be clade- or even organism-specific. A number of recent large-scale studies have lifted the number of miRNA loci known in human to 533 (Table 1) (15–17), around 60% of which are obviously conserved in mouse (miRBase release 10.0).
Table 1.
The number of published hairpin precursor and mature miRNA sequences in selected model organisms
| Hairpin precursor loci | Mature miR sequencesa | ||||
|---|---|---|---|---|---|
| Total number | Clustered ≤10 kb from another miRNA | Overlap annotated transcripts | Distinct forms | Experimentally verified | |
| Homo sapiens | 533 | 190 (36%) | 267 (50%) | 555 | 546 (98%) |
| Mus musculus | 442 | 199 (45%) | 174 (39%) | 461 | 455 (99%) |
| Danio rerio | 337 | 151 (34%) | 41 (12%) | 193 | 183 (95%) |
| Caenorhabditis elegans | 135 | 34 (25%) | 23 (17%) | 135 | 135 (100%) |
| Drosophila melanogaster | 93 | 34 (36%) | 36 (39%) | 88 | 85 (97%) |
| Arabidopsis thaliana | 184 | 19 (10%) | 16 (9%) | 199 | 199 (100%) |
| Populus trichocarpa | 215 | 42 (20%) | 9 (4%) | 215 | 55 (26%) |
amiR* sequences are excluded from the mature miRNA count.
miR and miR* sequences
The 5071 miRNA hairpin loci in the database express 4922 dominant mature miRNA (miR) products (Table 1). In many cases, deep sequencing technologies have detected large numbers of miR* sequences—biogenesis byproducts that are often detected at very low levels and are likely non-functional. Starting in miRBase release 10.0, mature miR and miR* sequences are better distinguished in the database, and distributed in separate release files. In many cases, mature miRNAs from both 5′ and 3′ arms of the hairpin precursor are frequently identified, suggesting that both may be functional, or there is insufficient data to determine the predominant product. Such miRNAs are given names of the form hsa-miR-140-5p and hsa-miR-140-3p, and both are retained in the miR set. Often, subsequent improved data allow one product to be chosen and annotated as the dominant miR. Recent data updates have occasionally caused the annotation of a miR and miR* pair to be reversed.
Variable ends
Increasingly deep and comprehensive cloning and sequencing studies identify many mature miRNAs with variable 3' (and, to a lesser extent, 5') ends [see for example (17)]. The miRNAs in the database currently represent the consensus of the most dominantly expressed sequence. As more data become available, the ends of mature miRNAs in the database will be adjusted to reflect the most up-to-date consensus information. We also aim to provide specific data on the distribution of ends in future releases. All changes in name and sequence between releases are specifically described in the diff file on the FTP site, along with all data from previous releases.
Experimental support
Usually the only available experimental data supports the mature miRNAs—hairpin precursors are very rarely experimentally validated. Rather, the precursors are the result of computational prediction of hairpin structures that include the mature miRNA. When a number of loci include the same mature miRNA, we cannot usually say with confidence which loci are actually expressed. In addition, the extents of the hairpins depicted in the database are somewhat arbitrary—the approximate extent of the predicted hairpin structure is shown. Formally, this includes the true precursor (the product of DROSHA cleavage) and a small amount of flanking sequence. Future developments will include the provision to retrieve the precursor with user-defined lengths of flanking sequence. About 3685 of 5922 mature miRNA products in the database are validated experimentally in the originating organism—the remainders are obvious homologues of validated miRNAs from a related species (Table 1). The ‘evidence’ field describes the origin of each sequence in the database.
miRBase::Targets
The miRBase::Targets database uses the miRanda algorithm (7) to predict targets in untranslated regions (UTRs) of 37 animal genomes from Ensembl (18). The quality of the predictions has recently benefited from significantly improved 3′UTR information, based on DITAG and 5′CAGE data, available from Ensembl. The number of human and mouse transcripts without an experimentally supported 3′UTR (for which we search a region 2 kb downstream) has therefore dropped significantly in the latest release (v5). A number of validated miR/target pairs are shown to have mismatches in the so-called ‘seed’ region (19). The miRBase/miRanda pipeline is therefore not constrained by the requirement for exact ‘seed’ matches. Recent papers have also highlighted the importance of secondary features for miRNA/target recognition, such as sequence accessibility, AU bias and UTR position (20,21). We intend to incorporate these features into the miRBase::Target prediction pipeline over the coming 12 months. In addition, links are provided to other target prediction sites and algorithms, and to the TarBase database of experimentally supported targets (22).
miRBase GENOMICS
Recently, we have focused on the provision of tools to distribute miRNA genomic information.
Genomic coordinates
Where an assembled genome sequence is available, coordinates of all miRNAs are provided: in summary tables for each organism and miRNA family, on each miRNA entry page, and for bulk download in GFF format. Links are provided from each coordinate to the appropriate genome browsers.
miRNA gene context
40–70% of vertebrate miRNAs appear to be expressed from introns of protein- and non-coding transcripts (Table 1) (23). In worms and flies, intronic miRNAs are less common (15% and 39%, respectively, in protein-coding genes), and only 5–10% of Arabidopsis miRNAs overlap annotated transcripts. For all animals with Ensembl-annotated genome assemblies, we provide a list of transcripts overlapping each miRNA, with overlap type (intron, exon and UTR), and sense (forward and reverse strands).
Clustered miRNAs
miRNAs are often clustered close together in the genome. This clustering has been suggested as evidence that >1 miRNA may be expressed from the same primary miRNA transcript (pri-miRNA). Furthermore, known ‘polycistronic’ miRNA transcripts are shown to be long: up to tens of kilobases in mammals. Over 40% of human miRNAs, over 30% of worm and fly miRNAs and only around 10% of Arabidopsis miRNAs are within 10 kb of another miRNA (Table 1). miRBase provides a list of clustered miRNAs on each applicable entry page. In addition, a new search facility allows the user to retrieve clusters of miRNAs in any organism separated by any choice of distance.
Genomic features
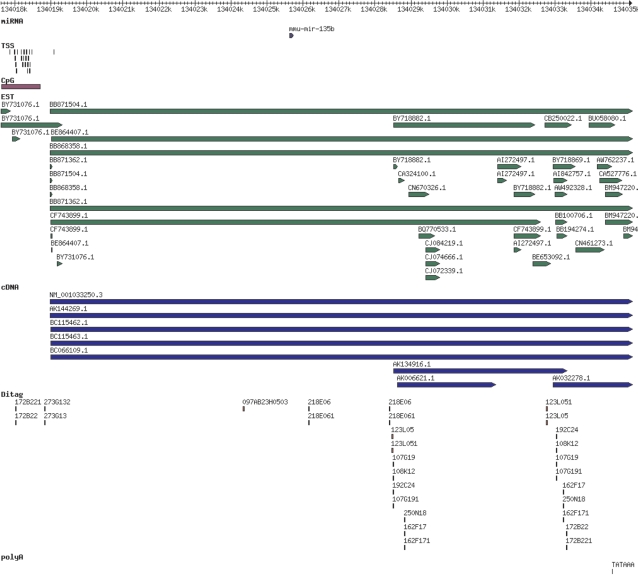
While the mapping of mature and hairpin miRNA sequences to assembled genomes is readily available in miRBase, the extents of only very few primary miRNA transcripts (pri-miRNA) are determined and annotated. For intronic miRNAs, the pri-miRNA is assumed to be the protein- (or non-)coding host transcript. Information about the extents of intergenic pri-miRNAs can be inferred from collective analysis of genomic features such as transcription start sites (TSS), CpG islands, EST and cDNA overlap, DITAG and 5′CAGE data, transcription factor binding sites (TFBS) and polyadenylation site predictions (polyA). A detailed analysis of these data suggest that pri-miRNA transcripts vary in length from a few hundreds of bases up to tens of kilobases (24). We have recently developed a tool to visualize the relative positions of these predictions and mappings with respect to annotated miRNA genes and clusters. Careful inspection of these data allows the prediction of the 5′ and 3′ boundaries of a significant number of putative pri-miRNAs. For example, Figure 1 shows TSSs, CpG island, ESTs, cDNAs, DITAG (172B22 and 172B221) and polyA site predictions surrounding mmu-mir-135b on mouse chromosome 1, which support a primary transcript of length around 15 kb with 5′ and 3′ ends ∼7–8 kb upstream and downstream of the miRNA. Links from each miRNA entry page provide a tabulated list of features overlapping flanking regions of the miRNA with their corresponding coordinates and scores, and a graphical view of the features present in the miRNA gene neighbourhood (as in Figure 1). These views are currently available for human, mouse, rat, worm and fly miRNAs, and will be extended to other organisms in the future. For human, mouse and rat genomes, TSSs are predicted using the Eponine-TSS software (25) at a threshold of 0.990. Drosophila TSS predictions, together with CpG islands, ESTs, cDNAs, repeats and DITAGs for all species are obtained from Ensembl. TFBSs in the flanking regions of human miRNAs are obtained from the conserved TFBS track of the UCSC genome browser (26). Other TFBS data are imported from the regulatory features track of Ensembl. PolyA signals are predicted in-house using the DNAFSMiner method (27) with a cutoff score of 0.6. The ‘Genomics’ section of the miRBase site allows the user to specify flanking and clustering distances, and the range of features desired.
Figure 1.
miRBase view of the distribution of genomic features around mmu-mir-135b on mouse chromosome 1, showing TSS, CpG island, EST, cDNA, DITAG (172B221 and 172B22) and polyA site support for a 15 kb primary transcript.
AVAILABILITY
miRBase is available on the web at http://microrna.sanger.ac.uk/. All data are available for download from the FTP site (ftp://ftp.sanger.ac.uk/pub/mirbase/) in a variety of formats including FASTA sequences and MYSQL relational database dumps.
ACKNOWLEDGEMENTS
S.G.-J. is funded by the University of Manchester. H.K.S. holds a GlaxoSmithKline postdoctoral fellowship, and work at the Sanger Institute is funded by the Wellcome Trust. Funding to pay the Open Access publication charges for this article was provided by the University of Manchester.
Conflict of interest statement. None declared.
REFERENCES
- 1.Molnár A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 2.Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol. Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell. Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 15.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, et al. The colorectal microRNAome. Proc. Natl Acad. Sci. USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berezikov E, van Tetering G, Verheul M, van de Belt J, van Laake L, Vos J, Verloop R, van de Wetering M, Guryev V, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16:1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 2006;13:849–845. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 20.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 21.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc. Natl Acad. Sci. USA. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Down TA, Hubbard TJ. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Han H, Li J, Wong L. DNAFSMiner: a web-based software toolbox to recognize two types of functional sites in DNA sequences. Bioinformatics. 2005;21:671–673. doi: 10.1093/bioinformatics/bth437. [DOI] [PubMed] [Google Scholar]